
Background
When someone asks you what animals looked like 30,000 years ago, you might describe a majestic scene like the picture above, or you might describe one of your favorite characters from Blue Sky Studio’s movie Ice Age. Now what if you were asked to describe the plants of the Ice Age? Many of us wouldn’t know where to start, and that’s okay because scientists are still discovering what vegetation was characteristic during the Ice Age. Let’s be honest, learning about a majestic mammoth or saber tooth tiger is way cooler than studying a puny arctic sedge, but knowledge of ancient plants hold important information related to studies on climate change and ancient animal habitat and diet.
With knowledge of a region’s past climate and plants, scientists can compare their composition to present day and see how climate change has shaped plant communities and make predictions about the future (Lascoux et al 2004). Plant fossils have been used to reconstruct the past by two methods 1) morphological identification by expert botanists and 2) genetic analysis. The fossil record can be compared to a catalog of ancient species…that has had a couple of pages ripped out. Unfortunately not every plant becomes a fossil, and not every fossil survives to present day. However, advances in the field of genetics have produced a new tool that can supplement fossil data: DNA barcoding.
Barcoding is a process that can identify a number of species from a sample by comparing a DNA region that is located in all of the species of interest (Li et al 2014). DNA barcoding is a way to detect species without having to physically see the species in the area or take a sample from the species; a clever way to account for elusive animals, or in our case, extinct plants. Animals and plants are constantly leaving bits of DNA, called environmental DNA, in the environment through shedding hair, root cells, pollen, and even their fecal matter. All that’s needed are samples of the environment, which can include soil, snow, feces and ice, and through DNA sequencing we can match these bits of environmental DNA and detect any species that was in the area! Many scientists have already recognized the value of barcoding as a tool to fill in the blanks left by the fossil record, and have begun sampling areas of preserved environmental DNA, such as ice cores, permafrost, cave and lake sediments (Bohmann et al 2014) (Gugerli et al 2005) (Pedersen et al 2013) (Willerslev et al 2014). Eske Willerslev is one of the notable pioneers in ancient, environmental DNA, and is known for his work with ancient DNA.

Central question
Willerslev’s research is focused on the extinction of ice age animals and the history of early European humans. He regularly travels to Siberia to collect ice cores and animal fossils from the Ice Age. However to fully understand the lives of ancient ice giants and the migration of early humans, we need a solid understanding of the environment that shaped their travel, homes and diets. From the samples he has collected, Willerslev has designed and led studies to recreate the history of Arctic vegetation to better understand the lives of extinct animals (2014). The study poses the initial question of Arctic plant composition during the last Ice Age, but also challenges the hypothesis made based purely from pollen and fossil records. Can DNA accurately assess the plant diversity with ancient DNA, and will it match pollen data?
Evidence
To answer these questions, Willerslev took over 200 samples from Arctic soils and began DNA barcoding. For plants, two regions of DNA were amplified, one from the chloroplast and one from the nuclear DNA, and three databases of plant genome sequences were used to increase accuracy of plant identification. Multiple loci analyses add more accuracy when trying to identify species. Multiple databases will also increase the accuracy of identification. Sometimes mistakes are made when entering a genome of nucleotide bases into a database, so it is good to cross reference between a couple databases. The results were also broken into three different time periods: Pre-Last Ice Age, During the Last Ice Age, and Post Last Ice Age (2014).
DNA gathered from the sediment contained plant species that are characteristic of two biomes: tundra and steppe environment. Only a few aquatic plant species were dated to the Post-Ice Age time period, giving a slight indication of a dry to wet shift after the Ice Age. A second set of data was needed to support this hypothesis, so Willerslev turned to nematodes.
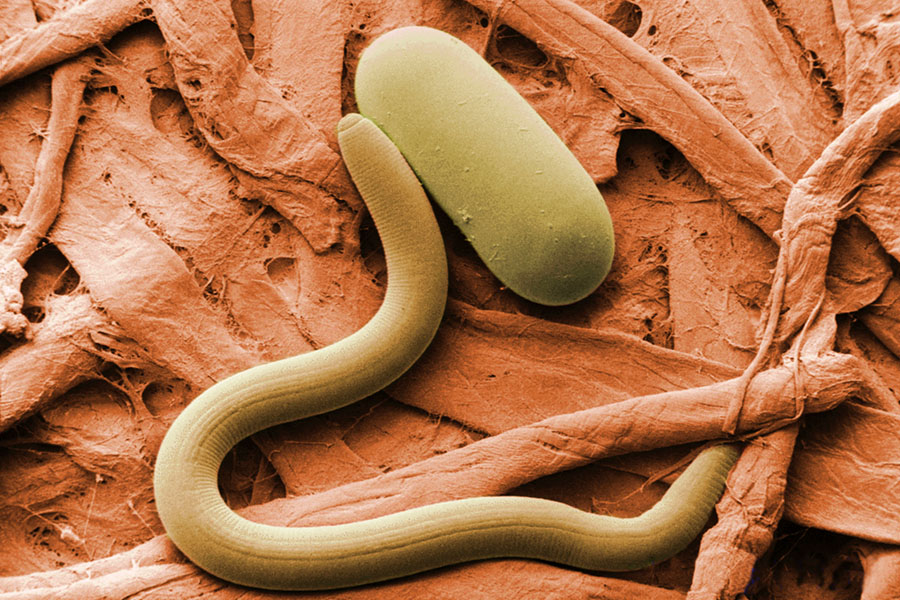
The ratio of nematode families is known to change with vegetation, moisture and climate (Yeates 2003), so DNA barcoding could be used once again to determine the relative amounts of nematodes. Dry-loving nematodes were found in all samples, whereas wet-loving nematodes were only found in samples dating after the last Ice Age. According to Willerslev, this matches previous studies that also indicate an Arctic shift from dry steppe or tundra to a moist tundra after the last Ice Age (2014).
While the data agree on a biome shift from dry steppe to wet tundra, the environmental DNA and pollen data were butting heads on the dominant plant type. The pollen data indicates that grasses were the dominant plant type Pre-Ice Age. Conversely, the environmental DNA indicated that the Pre-Ice Age time period was mostly populated by clovers and other wildflowers, with grasses accounting for only 20% of the diversity (Willerslev 2014). Clovers were also shown to be abundant in the other two time periods, but grasses only became the dominant plant type in the last 5,000 years, which is the plant usually associated with steppe/tundra environment in present day (2014).
To back up his environmental DNA, Willerslev tested whether mammoths and other ancient animals had diets mainly of clovers and wildflowers or grasses. They started by reanalyzing the sediment samples, this time amplifying a mitochondrial sequence found in all mammals, and then genetically tested stomach contents of mammoth and woolly rhinoceros, bison and horse specimens (2014). Willerslev’s team found that both the environmental and gut DNA was dominated by clovers. Clovers are rich in nutrients and more easily digested than grasses, which could explain how large mammals were able to maintain their size.
In conclusion, the nematodes, environmental DNA and glacial mammal gut contents all support the hypothesis of a dry steppe or tundra before and during the last Ice Age that was dominated by clovers. Mammals may have had an easier time maintaining their large size by having access to nutrient rich clovers. After the last Ice Age, the vegetation would have made a shift to a wet tundra or steppe environment that was still clover dominated, but eventually saw a decline in clovers and an abundance of grasses. I encourage you to look at more of Eske Willerslev’s research and interests on the Natural History Museum of Demark website: https://geogenetics.ku.dk/staff/?pure=en/persons/26558 .
Further reading
For more of Mauricio Antón’s depictions of ancient life, visit his website at https://mauricioanton.wordpress.com/ .
Interviews with Eske Willerslev about his research:
For more about DNA barcoding, here’s a review: DNA barcoding for ecologists by Alice Valentini et al. doi:10.1016/j.tree.2008.09.011
References
Bohmann, K., Evans, A., Gilbert, M.T.P., Carvalho, G.R., Creer, S., Knapp, M., Yu, D.W. & de Bruyn, M. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology & Evolution 29 (6), 358-366. doi: 10.1016/j.tree.2014.04.003
Core J. (2004). New soybean line offers strong resistance to nematodes. Agricultural Research Service: USDA <https://www.ars.usda.gov/is/pr/2004/040729.htm>.
Douka K, Higham T, Mastora E. (2015). Copenhagen sampling. PalaeoChron: Precision dating of the Palaeolithic :chronological mapping of the Middle and Upper Palaeolithic of Eurasia. <https://palaeochron-project.wix.com/palaeochron#!Copenhagen-sampling/cr1v/0CDA26CC-46E9-42A7-ABDE-403FDAEC5E19>.
Gronstal, A. (2009). Klondike holds clues to ancient environment. Live Science. Retrieved at https://www.livescience.com/5832-klondike-holds-clues-ancient-environment.html
Gugerli, F., Parducci, L. & Petit, R. J. (2005), Ancient plant DNA: review and prospects. New Phytologist, 166: 409—418. doi: 10.1111/j.1469-8137.2005.01360.x
Lascoux, M., Palmé, A. E., Cheddadi, R., & Latta, R. G. (2004). Impact of Ice Ages on the genetic structure of trees and shrubs. Philosophical Transactions of the Royal Society B: Biological Sciences, 359(1442), 197—207. doi:10.1098/rstb.2003.1390
Li, X., Yang, Y., Henry, R.J., Rossetto, M., Wang, Y. & Chen, S. (2014). Plant DNA barcoding: from gene to genome. Biological Reviews 90, 157-166. doi: 10.1111/brv.12104
Pedersen, M.W., Ginolhac, A., Orlando, L., Olsen, J., Andersen, K., Holm, J., Funder, S., Willerslev, E. & Kjaer, K.H. (2013). A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quaternary Science Reviews 75, 161-168. doi: 10.1016/j.quascirev.2013.06.006
Willerslev, E., Davison, J., Moora, M., Zobel, M., Coissac, E., Edwards, M. E., & … Binney, H. (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature506(7486), 47-51. doi: 10.1038/nature12921
Yeates, G.W. (2003). Nematodes as soil indicators: functional and biodiversity aspects. Biology and Fertility of Soils 37, 199—210. doi: 10.1007/s00374-003-0586-
