You probably know someone — or are someone — who says ‘____ drug just doesn’t work for me.’ or, ‘____ drug really messes me up’. Individual response to drug dosage is a pervasive confounding issue in health care. We know some of the pieces of the puzzle; age, metabolism, activity, and overall health are all factors contributing to individual drug response, but what if your guts have something to do with it, too? Bacteria present in the human gut make up what is called the ‘Human microbiome’, and it is the newest frontier in health research.
Background:
Prescribed drug use in some countries has increased by more than 250% in the last decade (WHO), so consequently the metabolism of drugs, along with their elimination and toxicity, has become a major area of concern for clinicians and researchers. We possess myriad enzymes to help our bodies absorb and convert any substance that comes from outside the human body , i.e. drugs (and food, of course) into usable, or bioavailable, forms. In the gut these enzymes transform drugs using either oxidation, reduction, or hydrolysis. However, there exists a whole host of microbes living within our gut that can metabolize drugs, too. These microbes can even modify the expression of enzymes produced by our own cells (Saad et. al., 2012).
According to a meta-analysis performed by Anubhav Das, Meenakshi Srinivasan, Tarini Shankar Ghosh, and Sharmilia S. Mande in 2015, there are significant correlations between the prevalence of these microbes and age, geographical location, and drug use. Many of the studies that have been recently performed to elucidate the function and effects of the human microbiome are carried out using a mouse model system, largely due to ethical and health concerns involved with using human subjects. However, there are limitations to using this research model, as mouse biology does not translate well to humans. There are differences in surface area and anatomy, and it has been found that there is an 85% dissimilarity in microbial composition between mice and humans (Ley et. al., 2005). Some researchers turned to lab culturing, but the majority of microbial species cannot be cultured using standard techniques (Amann et. al, 1995). For these reasons, Das et. al. turned to the field of metagenomics in their attempt to illuminate any relationships between geographic and age-specific variations in the drug metabolizing capabilities of the human gut microbiome.
*metagenomics: the study of genetic material recovered directly from environmental samples. In this case, the human gut (fecal samples).
Evidence:
Geographical differences
Using 397 publicly available metagenomic data sets from various age groups and 8 nationalities, the authors compared age, nationality, per capita drug use, and known drug metabolizing bacteria present in the samples. The authors identified 3 distinct clusters of microbe genera which produce similar enzymes but exhibit a range of versatility. Also within these groups, Das et. al. identified well-studied drug modifying enzymes (DMEs). The abundance of these DMEs are found to follow the same pattern as the diversity of microbe genera, establishing groups of low, intermediate, and high versatility microbe genera and DMEs.
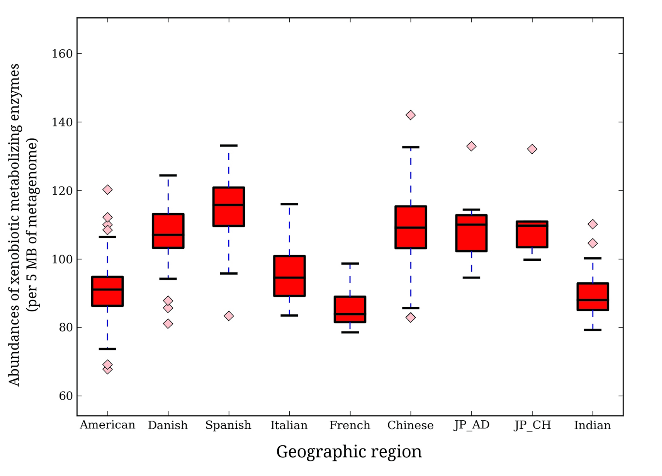
Spanish individuals had the highest abundance of metabolizing enzymes, followed by Japanese, Chinese, and Danish, with the Chinese having the highest degree of variation between individuals. Following the Danish were the Italian samples, then American, Indian, then French possessing the lowest metabolizing enzyme abundance. Their results suggest that individuals in the same geographic region have unique drug metabolizing capabilities compared to individuals in another location (Fig. 1). Furthermore, the types (at the genus level) of metabolizing enzymes present differed between geographic regions as well, with some geographic groups lacking bacteria that others possessed and vice versa.
Metabolizing Enzyme Abundance by Geographic Region
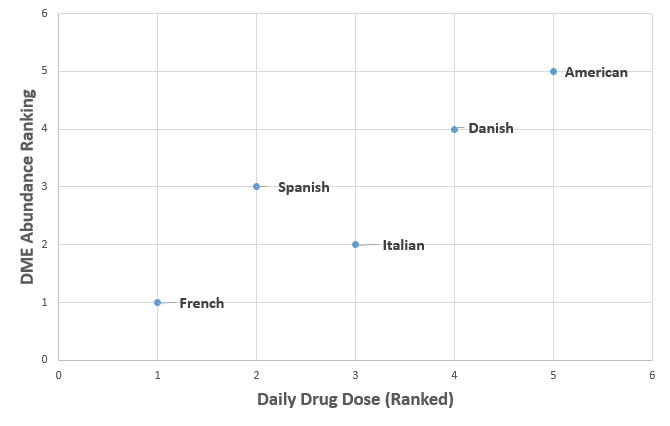
A positive relationship was observed between drug use and DME’s present in all populations except for the Italian cohort, indicating that the more drugs we take or foreign substances we ingest, the more drug metabolizing bacteria we harbor in our guts ( Fig. 2). Das et. al. looked at the specific abundance of enzymes known to have drug metabolizing capabilities (the DME’s), and they observed a geography-specific clustering by sample group. The Asian cohorts showed regional clustering, and the European groups had higher specific abundance of DME’s. To determine if these geography-specific trends correlated with drug use, the authors compared the abundance of metabolizing enzymes to the available daily drug dosage data, measured in drug units per day per capita.
Abundance of Drug Metabolizing Enzymes by Geographic Location
So what about age?
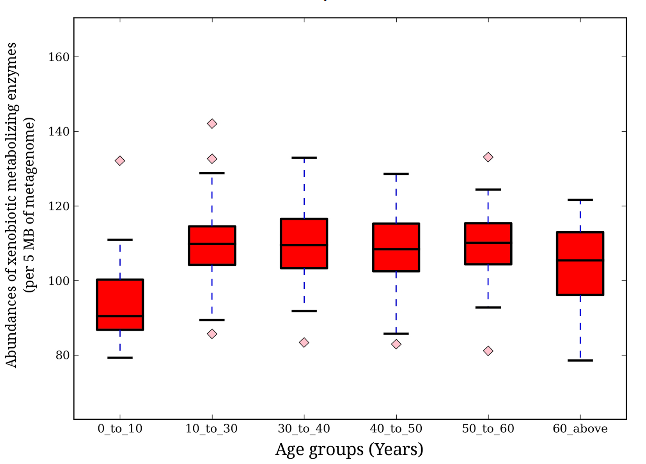
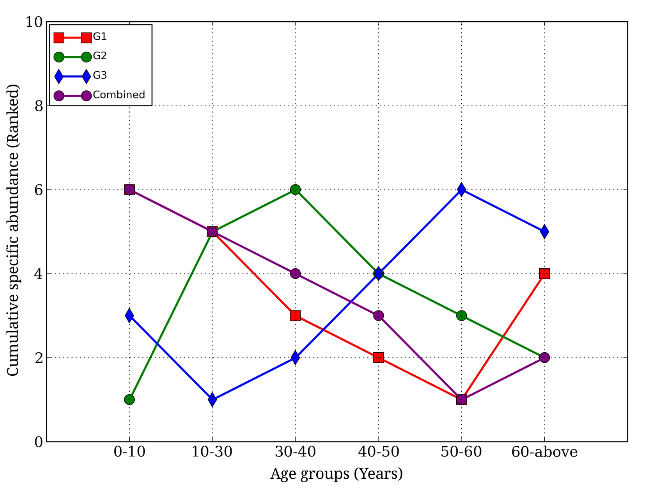
Do metabolizing enzymes present vary between age groups? The authors found that as a person ages, their microbiome changes to become more efficient at converting food and drug material into usable forms. When the samples were grouped by age; 0-10, 10-30, 30-40, 40-50, 50-60, and 60+ years, significant variations between age groups are apparent. The lowest numbers were found in the 0-10 age group, followed by an increase in abundance in the 10-30 age group, and similar amounts in the rest of the older age groups (Fig. 3).
Variation of Foreign Material Metabolizing Enzyme Abundance Across Age Groups
This conclusion was further supported when the authors examined the levels of DME’s across age groups. There are two phases of drug reactions in the gut. Phase I consists of oxidation, reduction, or hydrolysis performed by any number of enzymes. Phase II is conjugation, or binding, of the substrate to another organic molecule which makes the substance less reactive in order to be actively transported into the circulatory system. DME’s involved with Phase I reactions were more abundant in the younger age groups, while Phase II DME’s were present in higher amounts in the older age groups. This may be due to drug exposure increasing with age (Fig. 4). When diversity of the microbiome was examined by looking at foreign substance metabolizing enzymes and overall microbiome enzymes, it increased with age.
Specific Abundance of Low, Intermediate, and High Versatility Metabolizing Groups by Age
Questions
In this meta-analysis, the authors found significant differences in the metabolizing enzymes present between geographic populations and across age groups. It is not known for certain what factors they are due to. For example, diet has been indicated as a cause of the variation in microbiomes across populations (Schnorr et. al., 2014), and varies widely across ethnic groups and geographic locations. Future analysis could illuminate more of the factors contributing to variations in the metabolic microbiome across geographic locales. Drug use data was not age specific, but country specific. It is not likely that children in the 0-10 age group are exposed to many drugs, aside from an infant’s exposure via the mother. The results of the meta-analysis align with what most people would assume — that children don’t consume as many drugs as adults, therefore they do not possess as many drug metabolizing enzymes. What would explain the spike in the level of high variability metabolizing enzymes in the 60+ age group, though? I would be interested in a comparison of drug intake by age group to shed more light on the age/DME relationship.
Future studies could include RNA sequencing of the microbiome, and metabolomics analysis to further characterize the different bacteria present and their function in relation to drug metabolism. Longitudinal studies in which individuals are sampled from a young age into adulthood would provide a clear picture of changes related to age, drug intake, and geographical location.
Further reading
To read more about drug metabolism and the gut microbiome:
- Xenobiotics and the human gut microbiome: metatranscriptomics reveals the active players. Luke K. Urell, Rob Knight. Cell Metab. 2013 Mar 5; 17(3): 317—318. doi: 10.1016/j.cmet.2013.02.013
- Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Corinne Ferrier Maurice, Henry Joseph Haiser, Peter James Turnbaugh. Cell. 2013 Jan 17;152(1-2):39-50. doi: 10.1016/j.cell.2012.10.052. PMID: 23332745
For more on the differences in composition of the gut microbiome by geography and age groups:
