Background
Microorganisms are present all around us, but our interaction does not stop at that. There are millions of these microscopic creatures living in and on our bodies. While this may seem alarming at first, most of these microbes cause no harm, and in many cases play an important role in our health. Studies have shown that our microbiota, which are microscopic organisms that live in our bodies, have a wide range of functions from supporting our immune system (Mazmanian et al., 2008), to helping us stay free of infection (Fredricks et al., 2005). Scientists are trying to distinguish the range of what a healthy human microbial composition is. By establishing this range, the hope is to use the microbiome to detect and treat a variety of diseases (Rosenthal et al., 2011).
Every person has a microbial composition unique to them, which is called the microbiome. Each area of the body varies in what types of microorganisms are present. A study (Ying et al., 2013) focuses on the composition of microbes living on the skin, and what factors may play a role in the abundance and diversity of the microbiota between individuals, as well as between different places on the body. Their study concluded that diet, age, and living conditions each play a role in the variety and abundance of microbes living on the skin. The information gathered in studies like this one provide information to help us better understand the difference between bacterial communities. Once we know what causes the differences, we can dig even deeper and find out the mechanisms that act on the body and microbes to cause this change. This information can be used to help us understand the link between microbial presence and disease, as well as possibly manipulate the microbiome to treat certain diseases, or even prevent them before they start.
Central Goals
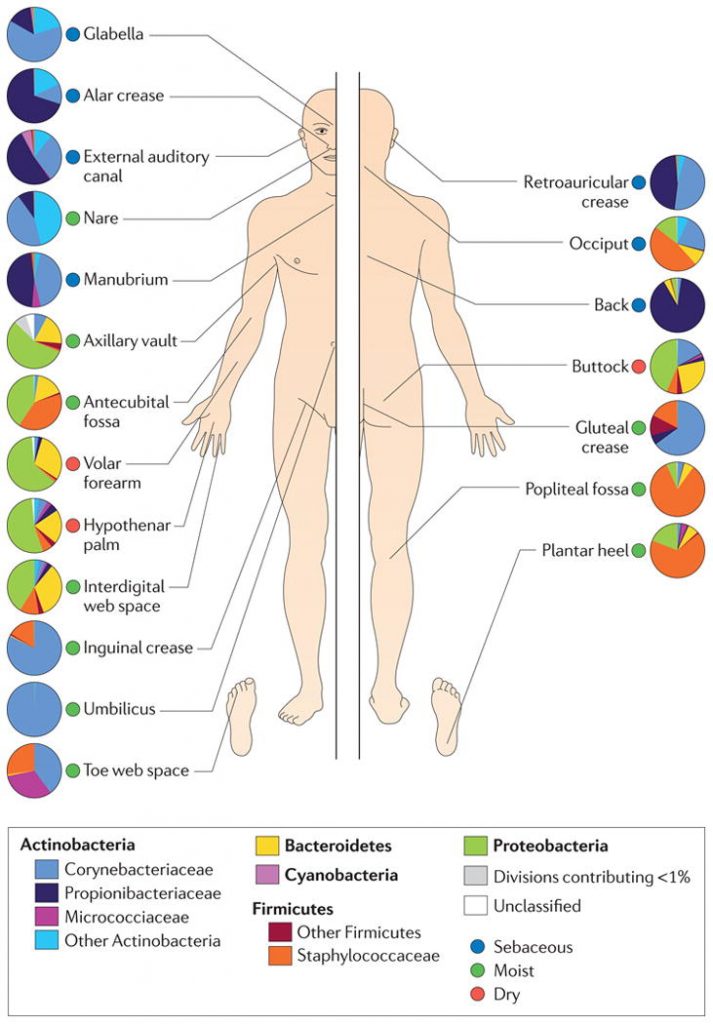
The make-up of microbial communities varies on different sites of the body (Grice, 2014). pH, temperature, moisture, and sebum influence which microbes are likely to populate an area.This study was conducted to determine if age, living conditions, and gender could account for variability of the skin microbiome. They also took into account skin conditions when assessing the results. It was hypothesized that each of these factors will play a role in the composition of the microbiota (Ying et al., 2013).
Evidence
71 individuals were grouped by age, gender, and living conditions. 36 of these individuals were living in urban areas and 35 were living in rural areas of Shanghai, China. They were grouped by age into three categories: adolescent (12-19), adults (25-35), and elderly (50-60). These individuals did not have any signs of dermatologic diseases, and did not use any type of antibiotic within the last 6 months. Seven sites on their skin were then sampled over a few days. DNA was extracted from these samples, a bacterial gene called 16s rRNA was amplified and the sequences were processed in a database in order to arrange the samples by genera, which are related groups. 16s rRNA is a well studied gene that is found in nearly all living organisms, and has many variable regions in which organisms can be distinguished from each other. These reasons make it a great target gene for studies.
Overall, the samples taken from adults showed not only more types of bacteria, but a greater number of bacteria as well. Differences that were observed between genders included a greater abundance of Propionibacterium on the forehead and hands from elderly males than females. Propionibacterium are commensal to humans and thrive near sebaceous and sweat glands.
A significant difference was found in the abundance of the genus Propionibacterium and Trabulsiella between individuals who lived in different areas. Urban dwellers had significantly more of these types of bacteria than rural dwellers. The opposite was true for the genus Corynebacterium, which was present in greater number in the rural individuals. Corynebacterium are another genera of commensal bacteria that live on the skin and often metabolize biotin and thiamin.
The largest difference was found between the urban and rural groups. Researchers set up a probability test that was able to distinguish whether an individual was from an urban or rural area solely based on the microbial composition of their skin.
Questions
Now that evidence has been gathered that shows differences in skin microbiota between age, gender, and living location; studies can be conducted to find out why. If the specific causes of these differences can be identified, they can be used to establish what a normal microbiota may look like for an individual. Understanding the mechanisms that affect microbial composition of the skin may also lead to possible therapies that can correct imbalances for conditions like atopic dermatitis (Myles, 2017). Studies could be conducted to assess why the differences in skin pH, moisture, or temperature affect the microbiome.
With so many variables between people, it may take a lot of careful planning to really be able to assess what factors affect the microbial diversity and why. Studies can also be conducted to better understand the balance between these differences and how it affects the individual’s health. By using articles such as The Skin Microbiome: Potential for Novel Diagnostic and Therapeutic Approaches to Cutaneous Disease (Grice, 2014), we can get an idea of what normal skin microbial composition may look like. If we can understand how to influence it, we may be able to use this knowledge to treat different diseases or encourage colonization of beneficial microbes to prevent disease.
It is likely that there will not be a specific microbial composition that is considered normal for everyone, but instead a range, much like vital signs that will fluctuate between individuals and over their lifetime. While there is evidence of correlation and association between microbes and diseases, there is still some mystery as to what mechanisms cause this.
Further Reading
- A study treated an area of eczema with bacteria found on healthy skin and lessened symptoms as a result. Bacteria therapy tested for common skin disease. (2018, May 15). Retrieved from https://www.nih.gov/news-events/nih-research-matters/bacteria-therapy-tested-common-skin-disease.
- This article highlights the variations in microbiota between different skin conditions. Grice, E. (2014). The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Seminars in Cutaneous Medicine and Surgery, 33(2), 98—103. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4425451
- This study treated mouse models that had eczema with transplanted healthy human skin microbiota. Myles, I. A. (2017). Transplantation of human skin microbiota in models of atopic dermatitis. Journal of Bacteriology & Parasitology, 08(05). doi: 10.4172/2155-9597-c1-037
- This video provides an explanation of the Human Microbiome Project and its future applications. Scientists re-define what’s healthy in newest analysis for human microbiome project. (2014, April 16). Retrieved from https://www.sciencedaily.com/releases/2014/04/140416133157.htm.
References
- Fredricks, D. N., Fiedler, T. L., & Marrazzo, J. M. (2005). Molecular Identification of Bacteria Associated with Bacterial Vaginosis. New England Journal of Medicine, 353(18), 1899—1911. doi: 10.1056/nejmoa043802
- Grice, E. (2014). The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Seminars in Cutaneous Medicine and Surgery, 33(2), 98—103. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4425451/
- Mazmanian, S. K., Round, J. L., & Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature, 453(7195), 620—625. doi: 10.1038/nature07008
- Myles, I. A. (2017). Transplantation of human skin microbiota in models of atopic dermatitis. Journal of Bacteriology & Parasitology, 08(05). doi: 10.4172/2155-9597-c1-037
- Rosenthal, M., Goldberg, D., Aiello, A., Larson, E., & Foxman, B. (2011). Skin microbiota: Microbial community structure and its potential association with health and disease. Infection, Genetics and Evolution, 11(5), 839—848. doi: 10.1016/j.meegid.2011.03.022
- Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., Gordon, J. I. (2008). A core gut microbiome in obese and lean twins. Nature, 457(7228), 480—484. doi: 10.1038/nature07540
- Ying, S., Zeng, D.-N., Chi, L., Tan, Y., Galzote, C., Cardona, C., Quan, Z.-X. (2015). The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. Plos One, 10(10). doi: 10.1371/journal.pone.0141842
