Background
In the 1940s Sir Alexander Fleming released the antibiotic Penicillin to the public transforming modern medicine forever. The emergence of antibiotics has had a profound impact on our lives, helping to increase our average life span from 56 to nearly 80 years (Ventola 2015). Antibiotics are an effective tool in fighting infection and have greatly reduced surgical complications. However, the flurry of excitement around these wonder drugs quickly went away. It was realized that the very microbes these drugs were supposed to be fighting were actually making them stronger and eventually became resistant to them.
Bacteria are able to trade genes through a process called horizontal gene transfer (HGT), almost like kids trading baseball cards. These genes are the instructions in our DNA for the proteins our cells need to survive and function. Some of these genes have the instructions for proteins that the bacteria can use to fight off antibiotic drugs and these are called antibiotic resistant genes (ARGs). The overuse and misuse of these drugs only spread ARGs. These genes are very diverse and have different effects within the cell; bacteria can accumulate multiple ARGs and become resistant to multiple antibiotics. On farms, animals are fed massive doses of antibiotics so that they can grow bigger and be healthier but the more they are exposed the better these bacteria become at withstanding antibiotics (Landers et al. 2012). The manure and urine these livestock produce then gets spread across the land and into underground water reservoirs which is then used to grow crops. The crops then pick up these same bacteria and become resistant themselves. As humans and other animals eat these livestock and plants the resistance of these bacteria spread into our bodies decreasing our effectiveness to antibiotics.
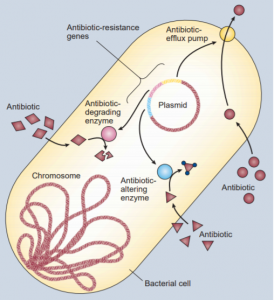
In 2006, the two most common hospital associated illnesses (sepsis and pneumonia) alone cost our healthcare system an additional $8 billion, doubled the average hospital stay to 12 days and were responsible for 50,000 deaths (Ventola 2015). Plus these numbers only account for the United States; the global cost is undoubtedly much more nerve wracking. Not to mention the burden this places on families and the wages lost from these extended infections. Up until now the solution to this spread of resistance has been to just make newer, stronger antibiotics. But economics, regulations and restrictions has made this harder and harder. The recent research into the topic of antibiotic resistance has been looking at how these microbes are able to protect themselves from these antibiotics and how prevalent they are in our environment.
Central Goals
A group of researchers (Li et al. 2015) from Hong Kong University and Michigan State University set out to look at the ARGs in the environment. From their sampled sites they aimed to determine what ARGs were present, similarities between them and if any of them could be used as indicators of ARG pollution.
Evidence
When Li et al (2015) looked at 50 different environments, from water, soils, bio-film to faeces, they found that antibiotic resistant genes were in all 50 environments. This means that there are bacteria that can survive our best efforts for fighting infection in all 50 of the environments they looked at. There was evidence of resistance to the 8 most commonly used antibiotics in the greatest abundance and distribution. This suggests that the more we use these drugs the more the resistance to them spreads, reducing their effectiveness and adding to the crisis. The most antibiotic resistant genes were found in human faeces, sewage treatment plants and the faeces/waste water of livestock farms.Not surprisingly, these are also the environments that come into contact with human and veterinary medicine the most. In the environments sampled, they showed that increasing microbe diversity also increases the number and different types of ARGs. Samples that have increased microbe diversity also have more ARGs and have more variation among them. That is the greater the diversity of a sample, the greater its ability to resist antibiotics. This is expected because when there are lots of different bacteria and each one of them has their own set of tools (their genes) which are specially adapted to their way of life which allowing them to live in some of the harshest environments Earth has to offer (Cary et al. 1997). Some of these tools are bound to be able to fight our medications. Since bacteria can trade their tools through HGT and they can reproduce really quickly, antibiotic resistance can spread within a population. One important thing to realize is that ARGs are natural and are a result of mutations to the genetic code. Microbes with these mutated genes, some of which have been found in areas that have never been in contact with antibiotics, can spread their resistant information on to other microbes (Davies and Davies 2010). This along with doses of antibiotics below the threshold needed to adequately fight bacterial infections is rapidly increasing the spread of antibiotic resistant bacteria.
Livestock farms harbored the most abundance of ARGs and thus had the most resistance. Farm ARG abundances were 10 to 1,000 times more concentrated than the samples from the soil and water. The high concentration of bacteria with antibiotic resistance in us and the food we eat is only making our fight against infection more difficult. It is not all bad news though. Despite the widespread use of one of our strongest and most common antibiotics, vancomycin, there were relatively low frequency of its resistant genes in all the environments (Li et at. 2015).
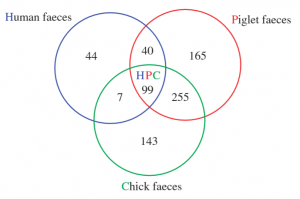
The researchers next compared the ARGs present in humans, pigs and chickens. They found that 85% of the ARGs in human were shared with young livestock, showing just how contagious these genes are (Fig. 2). When livestock become resistant from antibiotic overuse it is only a matter of time before humans acquire the same resistance. When all 50 environmental sites were looked at, patterns between the genera of microbes present; what ARGs they possessed and what antibiotics they were resistant to were found. Some genera of microbes were more likely to have specific categories of ARGs than others making them resistant to certain medications. Now comes the question – if a patient gets food poisoning caused by bacteria that is resistant to tetracycline, a common antibiotic then why not just use a different one? Well, Li et al. showed that microbes are able to harbor multiple ARGs at the same time – making them resistant to multiple medications. With all the different types of ARGs out there it can be difficult and time consuming to determine if a sample is antibiotic resistant. To make surveillance more feasible Li et al. purposes using two specific ARGs, tetM and aminoglycoside resistance protein, as indicators of antibiotic resistance. These two different types of ARGs were found to be indicators of others, so they may serve as a good indicator of overall antibiotic resistance.
Future Research
The goals of future research should be to find more cellular functions and proteins for future antibiotic drugs to target. On top of this one of the limitations of studies looking at the catalog of antibiotic resistance in an environment is that they are based on our current list of known ARGs which is by no means exhaustive. By focusing future research on looking for new ARGs we not only expand our list which increases the accuracy of studies like li et at. 2015 but this can also provide new strategies for antimicrobial medications to fight infection. Along with developing new antibiotics, scientists are exploring new, exciting classes of drugs. One of these exciting new frontiers is phage therapy which works by introducing a modified virus to an infected host. This virus is not like the flu or any other virus. It has been designed to fight only the infection causing bacteria. The virus has a special protein called Lysin which pokes a hole in the bacteria and kills it (Sulakvelidze et al. 2001). For this to be an effective treatment more of these phages need to be developed and tested. Modern medicine has moved past the age of antibiotics and the next treatment needs to be developed and introduced into hospitals.
Want to Learn More?
Check out these helpful links if you want to learn more about some of the topics mentioned above:
- Evolution of Bacteria Want to see just how long it takes bacteria to evolve and become resistant? If so check out this video based off an experiment at Harvard where they expose bacteria to different concentrations of antibiotic resistance and show just how fast bacteria are able to evolve against our medications.
- Horizontal Gene Transfer and Antibiotic Resistance Want to learn more about how microbes can transfer genes to one another? Watch this video to see how this process works and how it specifically applies to antibiotic resistance.
- Phage Therapy This video will give you a brief introduction into the future of phage therapy and what the potentials are as a clinical treatment.
References
- Cary, S. C., Cottrell, M. T., Stein, J. L., Camacho, F., & Desbruyères, D. (1997). Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Applied and Environmental Microbiology, 63(3), 1124—1130.
- Davies, J. (1996). Origins and evolution of antibiotic resistance. MicrobiologÃa (Madrid, Spain), Vol. 12, pp. 9—16. https://doi.org/10.1128/mmbr.00016-10
- Landers, T. F., Cohen, B., Wittum, T. E., & Larson, E. L. (2012). A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Reports, Vol. 127, pp. 4—22. https://doi.org/10.1177/003335491212700103
- Levy, S. B., & Bonnie, M. (2004). Antibacterial resistance worldwide: Causes, challenges and responses. Nature Medicine, Vol. 10, pp. S122—S129. https://doi.org/10.1038/nm1145
- Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. M., & Zhang, T. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME Journal, 9(11), 2490—2502. https://doi.org/10.1038/ismej.2015.59
- Sulakvelidze, A., Alavidze, Z., & Morris, J. G. (2001). Bacteriophage Therapy. Antimicrobial Agents and Chemotherapy, 45(3), 649—659. https://doi.org/10.1128/AAC.45.3.649-659.2001
- Ventola C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. P & T : a peer-reviewed journal for formulary management, 40(4), 277—283.
