Background:
Do you think humans could be a multi-planetary species? What would need to happen to make that dream a reality? These questions have spurred scientific researchers to develop projects that explore logistics of human survival in outer space, especially in an isolated and confined environment (ICE) over time. One such project is the Hawai’i Space Exploration Analog and Simulation (HI-SEAS). This is a NASA and University of Hawaii funded simulation that is set up to imitate a base camp on Mars or the Moon. Several crewed missions have been run at this “base camp” – the longest, Mission IV, had six crew members live there for a year. Mission IV included several scientific experiments amongst different disciplines such as psychology, botany, physiology and microbiology.

Microbiology is especially interesting because human beings have microbes, living on and inside them, that are critical to maintaining health. Microbes can be responsible for disease but also for healthy functioning of body systems such as the digestive, immune, and nervous systems (Young 2017). Since humans have a changed immune response in outer space it is a unique challenge to prevent risks of infection by maintaining a healthy human microbiome and minimizing detrimental microbes in the environment (Sonnenfeld 2005, Mermel 2012, Voorhies et al. 2019).
Some environmental microbes have been shown to survive extreme climates or thrive in isolated and confined environments (ICE) such as “technophiles” that can degrade common spacecraft materials like polymers and aluminum-magnesium alloys, “pyschrophiles” that can survive intensive cleaning procedures in the Jet Propulsion Laboratory at NASA or Deinococcus radiodurans that survived on the outside of a spacecraft in orbit for a year (Alekhova 2005, Danko et al. 2021, Ott et al. 2020). All confined settings have organisms that can survive yet it is still to be determined exactly how they are environmentally transmitted (Mora et al. 2016). A 2007 Mars mission simulation in Russia showed that hotspots for microbial growth are on humans and the areas they spend the most time (Schwender et al. 2017).
The HI-SEAS Mission IV provided another opportunity for researchers to try and map transmission patterns of microbes in an ICE. Researchers from the Medical University of Graz in Austria and the Space Life Sciences Lab at the University of Florida had crew members from Mission IV collect samples from their bodies and several spots in their environment every other week for a total of 111 microbe samples (Mahnert et al. 2021). The samples were then shipped to Europe when the mission was over for lab analysis. Although the HI-SEAS mission could not simulate an exact space basecamp with microgravity or radiation, they were still able to explore microbiota and human interactions in isolated and confined spaces to understand microbial movement patterns.
Central question:
How did the microbial communities shift and change over time, both on the crew and on the surfaces around them, during the HI-SEAS IV mission?
Evidence:
The researchers hypothesized that the number of microbes from each sampling location would start to look more similar over time (Mahnert et al. 2021). There were 6 human (or crew) sampling spots and 4 in the environment (bedroom desk, kitchen floor, common area desk, and toilet seat). They found that the overall microbial diversity and composition of biotic (skin) and abiotic (built environment) surfaces differs significantly. This result was found by analyzing each of the 111 samples and comparing it to the other samples. In microbial analysis this is done by asking two questions:
1) How many microbes are there?
2) How are the microbes balanced (is there evenness or do some species dominate others)?
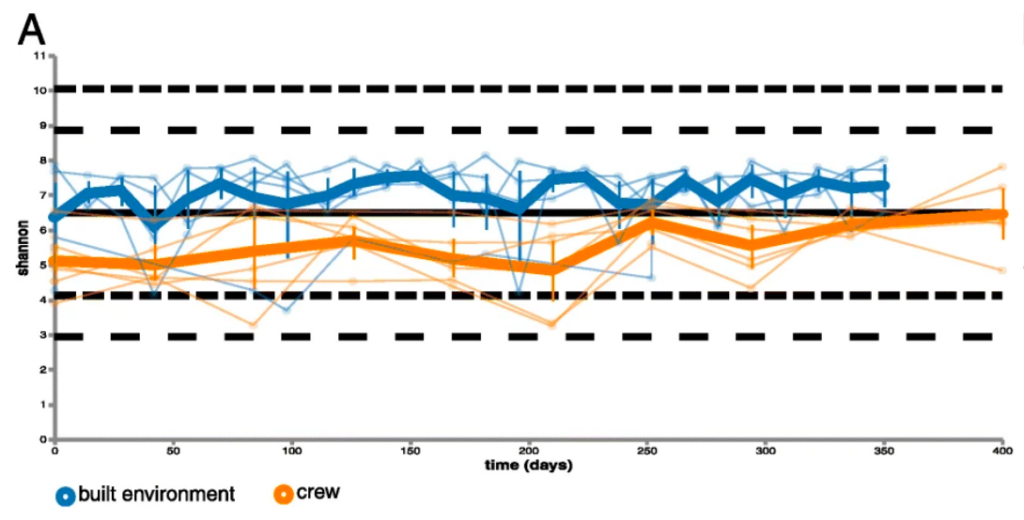
A statistical analysis, called the Shannon Diversity index, allows the researchers to answer those two questions with one number. Values in the Shannon index can range from one (if there is a single dominant species) to the total number of all species (if everyone present is equal). That number was then plotted on a graph to show the differences between the built environment samples and the crew member samples. Figure 1 shows us these results visually with a blue plot representing the built environment whose median is just above 7 and an orange plot representing the crew with a median just below 6.
They also found that microbial diversity on skin increased during the isolation period. This increase is represented in Figure 2, which graphically shows that while both sample diversities increased over time, the orange crew line increased more than the blue environment line. Using the Shannon diversity index, they found that skin diversity increased at a quicker pace than the built environment. As described earlier, a higher number means a greater number of species and balance of species that are present.
The researchers also wanted to know what kind of microorganisms were present at the different sampling sites. They answered this question by looking at each crew member and each built environment site separately and pooled together as “crew” and as “built” environment. They found that each surface was characterized by a specific set of microbial signatures, which can be predicted with high accuracy. “Microbial signatures” meaning that a sample could be matched to the sample site without knowing where it came from based on the microbial content. This is mainly identified by indicator species, which is an organism that occupies a specific environmental niche and whose presence and abundance can reflect the conditions of the sampling site (De Cáceres et al. 2010). For example, the microbe Kocuria was an indicator species for crew member A as seen in the red box in Figure 3 (Mahnert et al. 2021).
The researchers also found that the movement of microbes is highly dependent on personal interactions of the occupants of the confined space (Mahnert et al. 2021). This includes human to human interactions and human to non-living surface interactions. Unsurprisingly, crew members that interacted most often had the most similar microbial communities. Crew member F was noted as the most social crew member had also had the highest similarity to other crew members. The desk in the bedroom had a surface composition more similar to the crews’ communities than the samples over time from the kitchen floor. The researchers used a sophisticated statistical tool called SourceTracker2 to come to these conclusions (Knights et al. 2011).
While the variation amongst microbiomes of sampling sites evened out over time, it did take longer than expected due to a malfunctioning toilet. The crew members had to manually clean out the toilet, which spread Methanobrevibacter throughout the habitat. Unexpected incidents like these might take future base camp members by surprise so it is important to do further research to understand ways for humans to quickly restore a microbial community.
My questions:
How can crew members utilize current and developing technologies to make real time microbial composition analysis possible? This could look like providing a DNA sequencing toolkit for them with a MinION, which is a pocket sized device that a trained crew member can use to sequence DNA for immediate microbial monitoring immediately after a sample is collected (Tyler et al. 2018). It will be important for crews and researchers to establish how often sampling should be done to be efficient while still ensuring there are no microbial community disasters that go unchecked.
Another area to explore is how to use pre- and probiotics to proactively manage crew gut microbiomes? Prebiotics are essentially compounds in food that feed the microbes in the human microbiome while probiotics are live bacteria that can be put into your digestive tract on either end (Quigley et al. 2019) If crew members are able to use real time data to examine their microbiota, how could they use this information to maintain a beneficial community of microbes for health? The HI-SEAS IV experiment introduced some food related microbes such as yogurt and cheese bacteria without carefully looking at the data or designing a protocol with controls for these microbes. It would be interesting to see a study that controlled diet carefully by introducing stressors to the human microbiome and then seeing if the microbe composition could be returned to homeostasis through the digestive system.
A third area for researchers to study is antimicrobial resistance (AMR) genes. What AMR genes are present and what are the population fluxes? If resistance genes were to develop and proliferate in an isolated and confined environment (ICE), it could be dangerous for crew members who have a minimal amount of equipment and supplies to deal with potential outbreaks. AMR genes have the ability to spread in an ICE, particularly as microbes develop resistance to either cleaning materials, such as the commonly used quaternary ammonia, or antibiotics the crew may take. Because crews use quaternary solutions to sanitize the environment on the ISS, it is important to have a good understanding of what steps might be taken to mitigate an AMR disaster.
Further exploration:
Video
- A short video that describes the scientific paper this blog post is based on “Microbiome dynamics during the HI SEAS IV mission and implications for space travel”.
- The Commander of the HI-SEAS Mission IV gives a Ted talk about the crew’s experience.
- NASA video showing communication with the HI-SEAS crew as they navigate the landscape to find a new environmental monitoring instrument and learn to use it.
Written
- Blog post reviewing a paper that discusses how pro- and prebiotics can be used to help manage the human microbiome during space travel. It contains a link to the original article too.
- Website that overviews the private company SpaceX’s plan to travel to Mars.
- Biotechnology and Planetary Protection Group at Nasa discusses the main projects they are focused on.
References:
Alekhova, T. A., Aleksandrova, A. A., Novozhilova, T. Y., Lysak, L. V., Zagustina, N. A., & Bezborodov, A. M. (2005). Monitoring of microbial degraders in manned space stations. Applied Biochemistry and Microbiology, 41(4), 382-389. doi.org/10.1007/s10438-005-0065-x
Danko, D. C., Sierra, M. A., Benardini, J. N., Guan, L., Wood, J. M., Singh, N., … & Mason, C. E. (2021). A comprehensive metagenomics framework to characterize organisms relevant for planetary protection. Microbiome, 9(1), 1-15. doi.org/10.1186/s40168-021-01020-1
De Cáceres, M., Legendre, P., & Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos, 119(10), 1674-1684. doi.org/10.1111/j.1600-0706.2010.18334.x
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., … & Kelley, S. T. (2011). Bayesian community-wide culture-independent microbial source tracking. Nature methods, 8(9), 761-763. doi.org/10.1038/nmeth.1650
Mahnert, A., Verseux, C., Schwendner, P., Koskinen, K., Kumpitsch, C., Blohs, M., … & Moissl-Eichinger, C. (2021). Microbiome dynamics during the HI-SEAS IV mission, and implications for future crewed missions beyond Earth. Microbiome, 9(1), 1-21. doi.org/10.1186/s40168-020-00959-x
Mermel, L. A. (2013). Infection prevention and control during prolonged human space travel. Clinical infectious diseases, 56(1), 123-130. doi.org/10.1093/cid/cis861
Mora, M., Mahnert, A., Koskinen, K., Pausan, M. R., Oberauner-Wappis, L., Krause, R., … & Moissl-Eichinger, C. (2016). Microorganisms in confined habitats: microbial monitoring and control of intensive care units, operating rooms, cleanrooms and the International Space Station. Frontiers in microbiology, 7, 1573. doi.org/10.3389/fmicb.2016.01573
Ott, E., Kawaguchi, Y., Kölbl, D., Rabbow, E., Rettberg, P., Mora, M., … & Milojevic, T. (2020). Molecular repertoire of Deinococcus radiodurans after 1 year of exposure outside the International Space Station within the Tanpopo mission. Microbiome, 8(1), 1-16. doi.org/10.1186/s40168-020-00927-5
Quigley, E. M. (2019). Prebiotics and probiotics in digestive health. Clinical Gastroenterology and Hepatology, 17(2), 333-344. doi.org/10.1016/j.cgh.2018.09.028
Schwendner, P., Mahnert, A., Koskinen, K., Moissl-Eichinger, C., Barczyk, S., Wirth, R., … & Rettberg, P. (2017). Preparing for the crewed Mars journey: microbiota dynamics in the confined Mars500 habitat during simulated Mars flight and landing. Microbiome, 5(1), 1-23. doi.org/10.1186/s40168-017-0345-8
Sonnenfeld, G. (2005). The immune system in space, including Earth-based benefits of space-based research. Current pharmaceutical biotechnology, 6(4), 343-349. doi.org/10.2174/1389201054553699
Tyler, A. D., Mataseje, L., Urfano, C. J., Schmidt, L., Antonation, K. S., Mulvey, M. R., & Corbett, C. R. (2018). Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Scientific reports, 8(1), 1-12. doi.org/10.1038/s41598-018-29334-5
Voorhies, A.A., Ott, C.M., Mehta, S., Pierson, D.L., Crucian, B.E., Feiveson, A., Oubre, C.M., Torralba, M., Moncera, K., Zhang, Y. and Zurek, E., 2019. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Scientific reports, 9(1), pp.1-17. doi.org/10.1038/s41598-019-46303-8
Young, V. B. (2017). The role of the microbiome in human health and disease: an introduction for clinicians. Bmj, 356. doi.org/10.1136/bmj.j831
