The human genome is made up of over three billion base pairs, the building blocks of DNA, but only a small part of that is actually protein coding DNA. Transposable elements (TEs) are a type of non-coding DNA that makes up approximately half of the human genome, and since their discovery in the 1940s, they have carried a bad reputation. Often referred to as junk DNA or parasitic genes, TEs are a type of mobile genetic element capable of moving around or “jumping’ within the genome. There are two main types of TEs: retrotransposons, which copy themselves and then reinsert the copies back into the genome and DNA transposons, which cut themselves out of the genome and move to a different site (Figure 1). When TEs move around, they sometimes land in the middle of a gene or even another TE, which is often detrimental to the function of that gene or TE. The function and potential benefits of TEs have not always been clear, but research has slowly started to unravel their mysteries. A group of scientists at Yale, lead by Gunter Wagner, recently found that TEs may have been responsible for the evolution of pregnancy in mammals.

Mammalian pregnancy is an incredibly complex trait that arose many thousands of years ago, and it can be very difficult to understand how such traits evolved. Mammals are comprised of two main classes. Monotremes, including the platypus, which lay thin shelled eggs that hatch after a short period of time. And therians which can be further divided into marsupials, with short pregnancies that nurture their young in pouches and eutheirans, including humans, with prolonged pregnancies.
The first step that Gunter Wagner and his team completed in their mission to understand the evolution of mammalian pregnancy was to determine which genes are critically important to pregnancy. To do this, they sequenced the DNA found the lining of the uterus of several eutherian mammals, a marsupial, and a monotreme to find what genes were actually being coded and expressed in that tissue. They then combined this data with data of the same type from several more species of eutherian mammals as well as some non-mammal species including bird and amphibians. Overall, the group found almost 20,000 genes that were involved in pregnancy. These genes came from a total of 14 species and this allowed the scientists to track what lineages each gene came from.
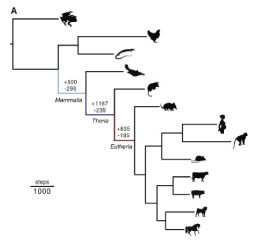
Once they found the genes of interest, they used them to construct a pylogenetic tree which shows how different species are evolutionarily related to each other (Figure 2). They were also able to use a method called parsimony to determine how many of the genes involved in pregnancy were gained and lost. Many of the genes that were gained were recruited from other organs in the body, such as those in the brain or gut (Figure 3), and functions of these gained genes often include, cell signaling, metabolic processes, and immune suppression. Maternal immunotolerance is important to pregnancy because the fetus has a unique immune system that the mother’s body would otherwise recognize as foreign and attack it. There were fewer genes that were lost in the evolution of mammalian pregnancy, and they often included genes involved in the formation of egg shells.

Recruited genes in mammalian pregnancy were more tissue specific than anciently expressed genes and that they were expressed at lower levels. Recruited genes are important for implantation of the egg and establishment of pregnancy because recruited genes were often much more misregulated than ancestral genes in women with unexplained infertility. In general, recruited genes had far more precise functions while ancestrally expressed genes carried out broader tasks.
After gathering information on what genes are actually involved in pregnancy, the group still had to find out how those genes were regulated. Genes in animals are regulated by other portions of DNA called promoters and enhancers that are located nearby the gene. These regulatory elements are responsible for controlling when a gene is activated, and how many protein copies it makes. The research team looked at the promoters and enhancers in each gene they found to be associated with pregnancy to see if there were any embedded sequences of known TEs. The results showed that ancient mammalian TEs were often integrated into these regulatory regions of DNA.
Another important component of gene regulation are proteins called transcription factors. These are proteins that bind to sections of DNA to help control when genes are expressed. This study found that transcription factors that regulate expression of genes involved in pregnancy often bind to sections of DNA that contain traces of ancient mammalian TEs.
In summary, several of the genes that are responsible for pregnancy in mammals originally had other functions in the body, and TEs played a role in the recruitment of these genes for pregnancy by coordinating and regulating them. TEs were able to do this by providing copies of the same DNA sequence nearby each gene in the enhancers, promoters, and transcription factors that can be stimulated by the same signals. This allows a group of genes to be controlled by a single signal, for example, the hormone progesterone. Overall, this study suggests that the evolution of pregnancy in mammals happened not by the slow development of a completely new set of genes, but by change in the regulation of groups of genes expressed in other systems of the body. Additionally, this study showed that the change in the regulation of genes was assisted by TEs. This mechanism of evolution is a way for complex systems to evolve much more rapidly than when single mutations within genes happen one by one.
Although TEs have often been thought of as undesirable junk, this study indicates that they are much more important than previously imagined. I would be interested to see the methods of this study repeated to find out if other systems of the body have evolved with the help of TEs. Other studies have showed that TEs may be responsible for the evolution of a range of traits from pigmentation patterns in fish to the vertebrate immune system, and a better understanding of TEs may answer important questions about cancer and aging. At first glance, TEs appear to be extremely disrupting, but further research may show that their less obvious functions are actually a crucial component in the evolutionary process.
Further Reading:
- https://www.nature.com/scitable/topicpage/transposons-the-jumping-genes-518
- https://www.nature.com/scitable/topicpage/transposons-or-jumping-genes-not-junk-dna-1211
References:
Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grutzner F, Bauersachs S, Graf A, Young SL, Lieb JD, DeMayo FJ, Feschotte C, Wagner GP. Ancient Transposable Elements Transformed the Uterine Regulatory Landscape and Transcriptome during the Evolution of Mammalian Pregnancy. Cell Rep. 2015. DOI: https://dx.doi.org/10.1016/j.celrep.2014.12.052
