Background
You’ve most likely had an infection that required treatment with antibiotics. At the time that you were prescribed antibiotics, you probably heard that familiar spiel about needing to take the entire course of antibiotics even if you feel better before finishing them. You might be wondering, why does it matter? Hopefully after reading this post you will be able to answer that question for yourself.
As you might imagine, bacteria developing the ability to survive antibiotic treatment is bad news for us. This means that over time, our commonly used antibiotics are becoming less effective at killing bacteria. As bacteria evolves more antibiotic resistance, we may no longer be able to treat common infections unless we come up with another alternative treatment. It is important that we try to delay the evolution of antibiotic resistance as much as we can to buy time to develop other effective treatments. So what are the driving forces of bacterial evolution and what can we do about it?
When they were first discovered, antibiotics were a revolutionary breakthrough. They extended life expectancies by preventing people from dying due to common bacterial infections (Hutchings et al, 2019). After the discovery was made, antibiotics were widely used not only to treat infections in medical settings, but also to prevent infections in agricultural settings especially among livestock (Manyi-Loh et al, 2018). This is partially still the case today. However, as time has passed, there is a new threat to the success of antibiotic treatments: bacterial evolution. Natural selection refers to genetic changes in a population of organisms over time that help that population thrive in their environment (Natural Geographic Society, 2019). Natural selection is a driving force of evolution. Like all forms of life, bacteria are constantly evolving. While antibiotics have proven very effective at killing bacteria, some bacteria can survive longer with antibiotic exposure and are less affected than other bacteria. Over time, the bacteria that can cope better with exposure to antibiotics will reproduce more than the bacteria that are killed. During the process of reproduction, bacteria pass their genes to future generations. As a result, the genes of the more successful bacteria will gradually become the most common genes in the bacterial population. In order to understand how much this will impact bacterial evolution, we must also understand the idea of a “bottleneck”. A bottleneck refers to an event in which the number of individuals in a population shrinks to a smaller than desirable size, resulting in reduced genetic diversity in the population (figure 1). There are a whole host of long-term consequences that will result depending on the severity of the bottleneck. This is because once genetic diversity is lost, it is hard to get it back. This can lead to more rare heritable diseases in the population, which can make it harder for the population to survive.
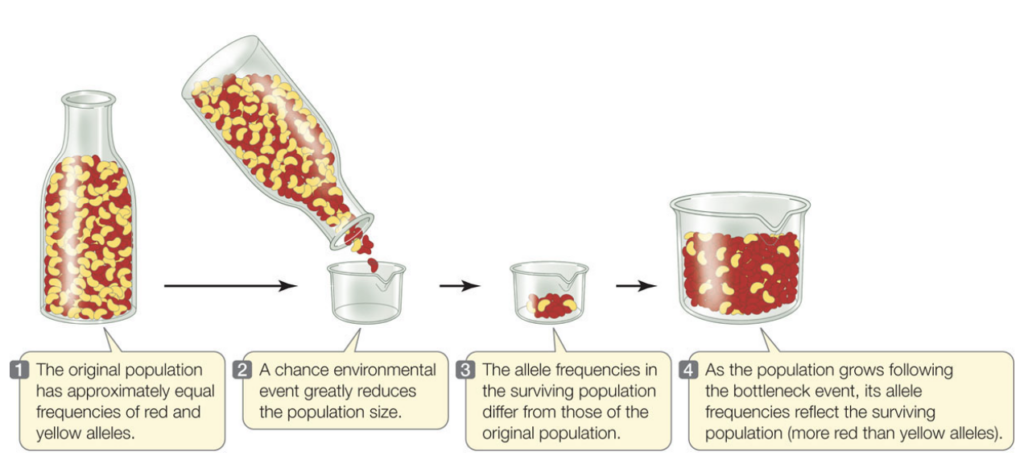
One recent study called “Bottleneck size and selection level reproducibly impact evolution of antibiotic resistance” explored the impacts of the size of a bottleneck in the bacterial population and the strength of antibiotic treatment the bacteria were exposed to in order to try and better understand the driving forces of bacterial evolution (Mahrt et al, 2021). The study treated both small and large bacterial populations with varying doses of antibiotics then observed how the populations changed over time and developed resistance to the antibiotics. Something important that the researchers discuss in the article is that it is not only the presence of antibiotics that dictate whether bacteria will evolve to have drug resistance. The experiment focused on how bacterial fitness and antibiotic resistance evolved in response to antibiotics.
Central Question
How does the size of a population bottleneck interact with varying strengths of antibiotic treatment to produce antibiotic resistance in future generations of bacteria?
Evidence
In order to determine whether the bacteria were becoming more fit (better at survival and reproduction in their environment), researchers compared the overall amount of bacteria present after each treatment to the overall number present at the same point in time in a control group that was not treated with any antibiotics (figure 2). Figure 2 shows the overall yields (amount of bacteria present) after antibiotic treatments. The researchers tested two different antibiotics in their experiment. To assess resistance in bacterial populations at the end of the experiment, the researchers analyzed their data using multiple techniques.
The researchers observed that populations with weak bottlenecks, that is larger populations, had higher fitness levels compared to those with strong bottlenecks at the end of the experiment. Regardless of which antibiotic was used or the dosage of the antibiotic, the researchers observed the most resistance in small populations with low doses of antibiotic (light blue) and large populations with high doses of antibiotics (red) (figure 3). However, all treatments did produce a higher level of antibiotic resistance in the population compared to the groups without antibiotic exposure. After further review of the data, the researchers concluded that the bacteria in the study were able to adapt the best when there were weak bottlenecks. Much of this was attributable to larger population sizes, minimizing the impact of random events and allowing natural selection to be the driving force of change in the populations. They also noted that all populations, regardless of the bottleneck strength, had higher fitness at the end of the experiment compared to the control group.
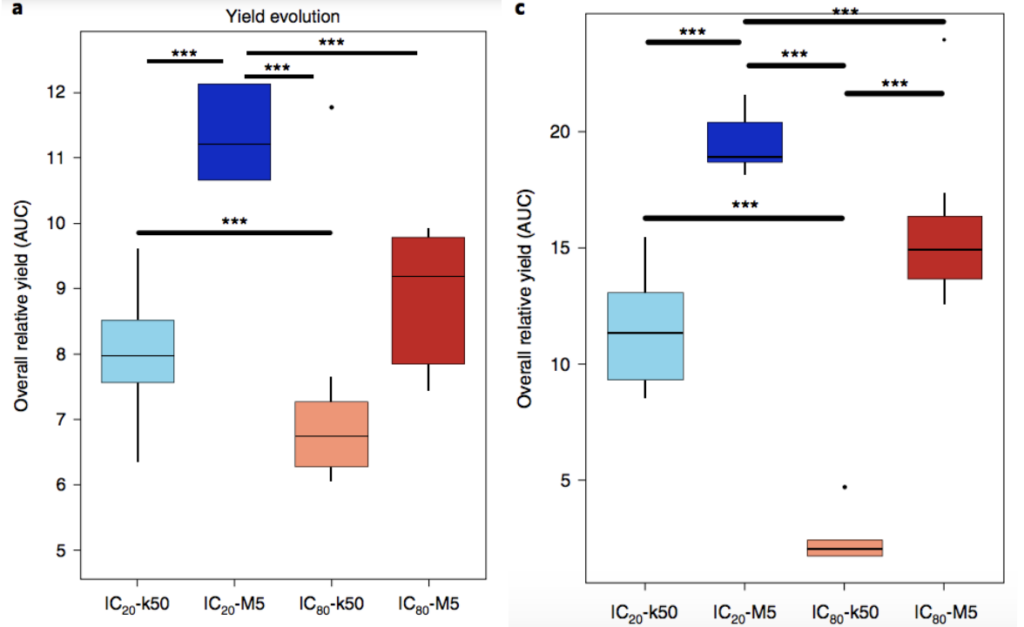
Consistent with expectations, when bacteria in large populations (weak bottleneck) are exposed to high doses of antibiotics, antibiotic resistance is favored and eventually becomes a common trait in the population. This reflects the predicted outcome of bacteria with higher fitness reproducing more frequently and passing their resistance genes to future generations. However, much to the surprise of researchers, low doses of antibiotics in small populations (strong bottleneck) have similar consequences. This is surprising because oftentimes, the effects of a strong bottleneck tend to override the benefit provided by a genetic mutation that aids the organism in survival. In other words, it is somewhat unlikely to see bacteria evolve antibiotic resistance in such a small population since it is super easy for beneficial mutations to be lost by random chance. Nonetheless, in the study when small populations were treated with low doses of antibiotics, they saw resistance become more common over time, indicating that the bacteria were evolving.
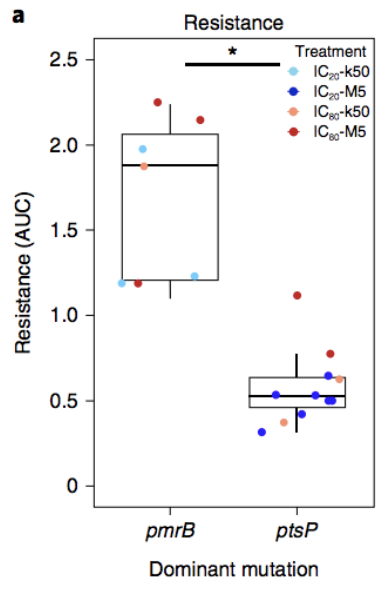
Considering the genetic changes that were responsible for changes observed in antibiotic resistance, there were two genes that the researchers paid attention to: pmrB and ptsP (figure 3). Figure 3 shows the difference in antibiotic resistance observed when each of the mutations was present. In this study, when pmrB was the main genetic change, there were higher levels of antibiotic resistance observed in the bacterial population. Analysis of the genetic material of the bacterial populations at the end of the experiment revealed that there were two genes with common variants when there were weak bottlenecks. However, in strong bottlenecks, there were more genes with common distinct variants. They observed that gene variant frequencies took longer to emerge with weaker antibiotic treatments compared to stronger ones. Specifically, new variants in the weak-treatment strong-bottleneck bacterial population did not become detectable until between 7 and 9 generations. However, the strong-treatment weak-bottleneck bacterial population had common new variants by the 3rd generation.
Future Research
Moving forward, this study offers meaningful insights into the development of antibiotic resistance. While the results of this experiment were observed in a lab environment, the experiments conducted do simulate real-world scenarios. For instance, we often hear about how antibiotic use in agriculture is a big issue for antibiotic resistance, and probably more impactful for the evolution of antibiotic resistance than medically prescribed antibiotics for active infections. Antibiotics were often used in agriculture as a preventative measure as opposed to a treatment for infections, called prophylactic use, until the practice was restricted in the US in 2017 (CDC, 2021). However, these restrictions did not completely ban the use of antibiotics, rather farmers now must gain approval from a veterinarian to utilize certain antibiotics. The new rules have simply restricted use of antibiotics that are “medically important” so they can only be used for “treatment, prevention, and control” of a specific disease (CDC, 2021). Antibiotic use in agriculture can be hugely problematic because it exposes a huge population of bacteria from a wide range of species to antibiotics and accelerates the shift toward all bacteria evolving resistance. This type of scenario would be very similar to what the researchers observed in their study with a large population being exposed to high doses of antibiotics. The study demonstrated that this type of scenario will consistently produce antibiotic resistance as a common trait in the population. More research on this specific scenario will provide a better understanding of how our commercial agriculture industry might specifically be driving antibiotic resistance trends. It is good that restrictions have been implemented for some antibiotics. By doing more research, hopefully we can understand more effective ways to use antibiotics in the short-term to slow the bacterial evolution process.
Considering antibiotic use in smaller populations of bacteria, it would be a good idea to do more research on how we can use alternative antibacterial treatments to handle common infections in people. There is promising research on how bacteriophages may help offer a long-term solution to antibiotic resistance. Bacteriophages are viruses that do not adversely impact human health, and specifically target and kill bacteria (Kasman and Porter, 2021). They are one of the main ways that nature keeps bacterial populations in check. Years ago there was more research being conducted on bacteriophages, but the research was largely abandoned with the advent of modern antibiotics. As we begin to realize that it is only a matter of time until most bacteria evolve to withstand our current treatments, we need to focus our research efforts on alternative options so lives are not unnecessarily lost. Preliminary research and knowledge about using bacteriophages as an alternative to antibiotics gives us some basic ideas about advantages and disadvantages. The advantages are pretty straightforward: bacteriophages are very specific and do not have negative effects on the host (such as imbalances and additional infections that result from antibiotic use), development of new phages takes less time and money compared to traditional antibiotics, and lastly phages can cross the blood-brain barrier which may allow them to more effectively treat infection of the brain or spinal cord (Romero-Calle et al, 2019). Most of the disadvantages have to do with knowledge gaps related to success of bacteriophages for treating bacterial infections in a clinical setting and little existing guidance to get a treatment of this nature approved by the FDA (Romero-Calle et al, 2019). The main functional challenge using phages is that our own immune systems may reduce the effectiveness of bacteriophages. Our own immune systems could recognize bacteriophages as viruses and kill them before they have a chance to put a dent in fighting the infection they were administered to treat (Romero-Calle et al, 2019). To combat this issue we could breed phages to avoid the immune system or we could manipulate dosages of phages given to patients (Romero-Calle et al, 2019). These challenges outline the main areas future research efforts should focus on in order to develop bacteriophages that would be viable treatment alternatives for antibiotics.
Further Reading
- This article provides an overview of the history of bacteriophage therapy as a potential alternative to traditional antibiotic treatments.
- This article from Vox discusses the current situation with antibiotic resistance and why it is problematic.
- This article discusses use of antibiotics in farming and current usage trends.
References
- Hillis, D. M. (2015). Mutation, Selection, Gene Flow, Genetic Drift, and Nonrandom Mating Result in Evolution. MacMillan HigherEd. Retrieved April 11, 2022, from https://www.macmillanhighered.com/BrainHoney/Resource/6716/digital_first_content/trunk/test/hillis2e/hillis2e_ch15_3.html
- Hutchings, Matthew I, et al. “Antibiotics: Past, Present and Future.” Current Opinion in Microbiology, vol. 51, 2019, pp. 72–80., https://doi.org/10.1016/j.mib.2019.10.008.
- Kasman, Laura M. “Bacteriophages.” StatPearls [Internet]., U.S. National Library of Medicine, 28 Sept. 2021, https://www.ncbi.nlm.nih.gov/books/NBK493185/.
- Mahrt, Niels, et al. “Bottleneck Size and Selection Level Reproducibly Impact Evolution of Antibiotic Resistance.” Nature Ecology & Evolution, vol. 5, no. 9, 2021, pp. 1233–1242., https://doi.org/10.1038/s41559-021-01511-2.
- Manyi-Loh, Christy, et al. “Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications.” Molecules, vol. 23, no. 4, 2018, p. 795., https://doi.org/10.3390/molecules23040795.
- National Geographic Society. “Natural Selection.” National Geographic Society, 7 Sept. 2019, https://www.nationalgeographic.org/encyclopedia/natural-selection/.
- Romero-Calle, Danitza, et al. “Bacteriophages as Alternatives to Antibiotics in Clinical Care.” Antibiotics (Basel, Switzerland), MDPI, 4 Sept. 2019, https://doi.org/10.3390/antibiotics8030138
- “UK Risks Falling behind on Reducing Farm Antibiotics after EU Ban.” The Guardian, Guardian News and Media, 28 Jan. 2022, https://www.theguardian.com/environment/2022/jan/28/uk-risks-falling-behind-on-reducing-farm-antibiotics-after-eu-ban.
- “Where Resistance Spreads: Food Supply.” Centers for Disease Control and Prevention, Centers for Disease Control and Prevention, 22 Nov. 2021, https://www.cdc.gov/drugresistance/food.html.
