
Background:
To the general public, the idea that there are tiny organisms living all around (and inside) of us might be a scary concept. Naturally, if all the news you get on a regular basis is concerning the totally-terrifying E. Coli that can give you food poisoning, or that fiendish-foe influenza–it’s not surprising that people often have negative reactions to the term “microbe.’ The reality is that we’re mostly made up of microbes, we encounter them every day, and most of the time they’re harmless or even beneficial! In fact, we often use microbes to ferment sugars so we can make things like yogurt and bread, and just as we use these microbes for our own benefit–plants can do the same!
Most information on beneficial plant microbes concerns soil microbes–specifically, mycorrhizae, which are fungi that provide the plant with increased nutrient and water absorption in return for some of the carbohydrates it makes during photosynthesis. Mycorrhizae and other soil microbes associated with plant roots are part of what is known as the rhizosphere–the small area that is influenced by and surrounds the roots of the plant (Mendes et al. 2013).
While we possess a wealth of information regarding the rhizosphere, the phyllosphere (the above-ground/aerial parts of the plant that can be colonized by microbes, known as epiphytes) is not as well-understood (Lindow and Brandl 2003). This might surprise you if you think about just how many plants there are in the world, and the magnitude of their combined surface areas!
But hark–do not despair! Phyllospheric studies (specifically those which are culture-independent, revealing previously hidden diversity due to the fact that we can only culture around 1% of microbes) have been progressing (Smalla 2004), (Muller and Ruppel 2013)! We know that the composition of the phyllosphere is different from that of the rhizosphere and the vast majority of the phyllosphere is made up of bacteria (Lindow and Brandl 2003). We also know that more pigmented bacteria colonize the phyllosphere than the rhizosphere–presumably due to the influence of solar radiation–and that common rhizosphere colonizers can fail to become established on the phyllosphere (Lindow and Brandl 2003).
It is also known that epiphytic bacterial populations differ in size among and within plants of the same species, as well as in close proximity, over short time scales, and over the growing season (Lindow and Brandl 2003). The species of plant itself also plays a role in determining epiphyte community composition, as different plants have different size leaves (ie: different carrying capacities), different levels of leaf-waxiness, and a whole range of other factors that can influence epiphyte community composition (Whipps et al. 2008).
Research GoalS:
Recently, researchers in Norway conducted an experiment to elucidate how the bacterial community composition on leafy greens develops over time (Dees et al. 2014). They believed there would be a change in the epiphyte composition of the phyllosphere due to a decline in nutrient supply following leaf maturation, as well as weather effects and/or the selection of specific epiphytes by different types of leafy greens (lettuce mainly). Specifically, they wanted to determine the change in epiphyte community structure throughout the growing season (April-September) for two kinds of leafy greens.
Evidence:
The researchers concluded that bacterial richness in lettuce was significantly greater 3 weeks after planting rather than at harvest (Dees et al. 2014). They came to this conclusion due to a significantly (P = 0.002) higher number of operational taxonomic units (OTUs) from lettuce samples collected at three weeks after planting than from samples collected at harvest (as can be seen in Fig 1; where the bars represent the 95% confidence intervals).
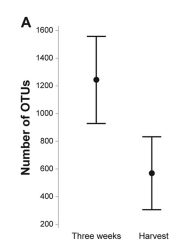
You might be wondering: ‘why the heck does more OTUs equal more bacterial richness?’
Well, in this study, the authors were using genetic sequencing as a way of studying the bacterial epiphytes associated with the phyllosphere of these leafy greens. As you may (or may not) know, all living organisms possess deoxyribonucleic acid (DNA), and by comparing DNA sequences we are able to determine how closely related species are to one another–the science of doing so is referred to as phylogenetics. In phylogenetics, OTUs are made by grouping sequences or groups of sequences that are most similar to each other (Van de peer 2009). So, having a lot of OTUs means you have many sequences that aren’t similar to one another–which gives us a decent measure of diversity because it means there were many DNA segments from organisms that weren’t closely related. Other studies have also shown that younger leaves are composed of a greater number of taxa than those of more mature leaves, so this finding isn’t entirely surprising (Lindow and Brandl 2003); however, the authors did also note that the finding wasn’t consistent in all of the samples.
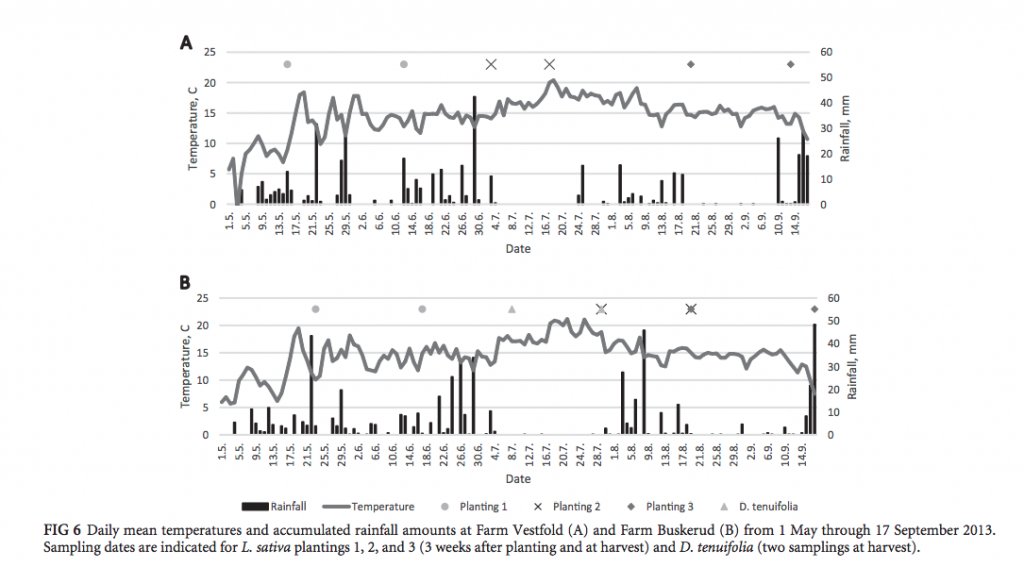
Specifically, they mention that the second planting (at both locations) possessed similar richness at 3 weeks and at harvest, as well as similar microbial profiles. They suggest that this may partly be due to the fact that they observed a higher temperature (at both locations) during harvesting of the second planting, than they observed for the harvesting of planting 1 or 3 (as can be seen in Fig. 2).
The authors statistically evaluated the OTUs of the lettuce samples to determine main bacterial distribution at 3 weeks after planting and at harvest, and found that the most abundant phyla at 3 weeks were different from the most abundant phylum found at harvest. If you take a second here and think back to biology classes and how living things are classified based on their taxonomic rank (kingdom, phylum, class, order, family, genus, species)–this is a striking finding because it essentially means that the bacterial communities present on the young 3 week old lettuce were completely different than those on the lettuce at harvest.
My Questions:
Most of the questions I have after reading this paper mainly regard the methods that were employed in the study–had I been conducting this study I would have restructured some of the methodologies employed. For example, the two farms used different water sources, had different methods of planting (bare-soil vs. plastic covered beds), different planting dates, etc. I would have liked to have seen more time points in this study, as well as synchronized planting, sampling, and harvesting days. In this regard, I suppose my questions concern how these different variables affect the phyllosphere communities, and by what degree.
Why didn’t you synchronize the planting (and sampling ) dates between the farms? What was the reasoning behind the time-points chosen/why not more?
I believe taking a look into the different possible sources of inoculation (water, soil, sampling protocol, sanitization protocols, etc) could provide an interesting glimpse into phyllosphere ecology. Additionally, the authors of this paper stated that one of the reasons for conducting this study was to determine the potential for human pathogens to be spread through leafy greens; however, the only mention of pathogens made in the paper was to state that they did not find any trace of E. Coli or Salmonella. I would go about asking them why they didn’t do any further investigations into the matter. I would also be interested in investigating what effect, if any, can be seen on plant development after they’ve been inoculated with certain human pathogens.
Further Reading:
For a short yet excellent overview of research that has been conducted on phyllosphere microbiology, I’ll refer you to The Microbiology of the Phyllosphere by Steven E. Lindow, and Maria T. Brandl (2003).
doi.org/10.1128/AEM.69.4.1875-1883.2003
If you’re interested in reading further about culture-independent advancements that have been made in phyllosphere microbiology, I recommend reading Progress in cultivation-independent phyllosphere microbiology, by Thomas Muller and Silke Ruppel (2014).
doi.org/10.1111/1574-6941.12198
A superb article concerning the diversity of phyllosphere microbiology and its relation to plant genotype is Phyllosphere microbiology with special reference to diversity and plant genotype, by J.M. Whipps, P. Hand, D. Pink and G.D. Bending (2008).
doi.org/10.1111/j.1365-2672.2008.03906.x
References:
- About the Human Microbiome. (n.d.). Retrieved from https://hmpdacc.org/hmp/overview/
- Dees, M. W., Lysøe, E., Nordskog, B., & Brurberg, M. B. (2014). Bacterial Communities Associated with Surfaces of Leafy Greens: Shift in Composition and Decrease in Richness over Time. Applied and Environmental Microbiology,81(4), 1530-1539. doi:10.1128/aem.03470-14
- Lindow, S., Brandl, M. (2003). Phyllosphere microbiology: A perspective. Microbial Ecology of Aerial Plant Surfaces,1-20. doi:10.1079/9781845930615.0001
- Microbes and food — Producers. (n.d.). Retrieved from https://microbiologyonline.org/about-microbiology/microbes-and-food/producers
- Mendes, R., Garbeva, P., & Raaijmakers, J. M. (2013). The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews,37(5), 634-663. doi:10.1111/1574-6976.12028
- Müller, T., & Ruppel, S. (2013). Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiology Ecology,87(1), 2-17. doi:10.1111/1574-6941.12198
- Plants and Mycorrhizae. (n.d.). Retrieved from https://sweetgum.nybg.org/science/glossary/glossary_details.php?irn=1074
- Smalla K. 2004. Culture-Independent Microbiology, p 88-99. In Bull A (ed), Microbial Diversity and Bioprospecting. ASM Press, Washington, DC. doi: 10.1128/9781555817770.ch9
- Successful soil biological management with beneficial microorganisms. (n.d.). Retrieved from https://www.fao.org/agriculture/crops/thematic-sitemap/theme/spi/soil-biodiversity/case-studies/soil-biological-management-with-beneficial-microorganisms/en/
- Taxonomy. (n.d.). Retrieved from https://basicbiology.net/biology-101/taxonomy
- Van de Peer, Y. (2009). Phylogenetic inference based on distance methods: theory. In P. Lemey, M. Salemi, & A.-M. Vandamme (Eds.), The phylogenetic handbook’¯: a practical approach to phylogenetic analysis and hypothesis testing (pp. 142—180). Cambridge, UK: Cambridge University Press.
- What is DNA? – Genetics Home Reference – NIH. (n.d.). Retrieved from https://ghr.nlm.nih.gov/primer/basics/dna
- Whipps, J., Hand, P., Pink, D., & Bending, G. (2008). Phyllosphere microbiology with special reference to diversity and plant genotype. Journal of Applied Microbiology,105(6), 1744-1755. doi:10.1111/j.1365-2672.2008.03906.x
