Background
Genetics and the environment; how do these interact? Do they always interact, or do genetics sometimes overrule characteristics learned from our environment? The question of nature, generally thought of to be our genetic make-up, versus nurture, the environments we’re exposed to in our developmental years, has been the topic of debate by scientists and philosophers for centuries. Yet, the definitive answer still frustratingly eludes us. Some things, like the number of limbs we’re born with, are entirely decided by genetic factors. Other things, like many of our behaviors, rely on an interaction between genetics and developmental environment.
The answer, it would seem, is highly dependant upon the context of the question. And the human salivary microbiome, the assemblage of microbes and their associated genomes and metabolites contained in the various niches of the mouth, presents us with a unique opportunity to investigate the impact our genetics and our environment could have on the organisms contained within our bodies and their effects on our day to day lives.
The Question
The research of Shaw et al. (2017) sought to investigate, quantify, and compare the effects of host genetic similarities, and shared host environment on the composition of the salivary microbiome. To do this, the researchers took advantage of a recently genotyped large Ashkenazi Jewish family (known as Ashkenazim) ( Zhernakova et al. 2015, Costa et al. 2013). Because of the close relatedness of the Ashkenazim, and their strict cultural practices, the researchers were able to control for and test the effects of human genetic relatedness and shared household environment. The researchers took salivary microbiome samples and described the microbial communities by targeting the 16S ribosomal RNA gene, the primary way we can characterize different groups of microbes in the microbiome.
The Research
Shaw et al. found that multiple factors, including shared household status and age, produced statistically significant differences between individuals. The authors used several powerful statistical tests, called Analyses of Variance (ANOVA), to analyze the amounts of variation between individuals’ oral microbiomes that can be explained by genetic and environmental factors. ANOVAs look at many different variables and determine, within a statistically acceptable margin of 5%, the amount of variation in the tested subject (the oral microbiome) that can be explained by the many different input variables. The authors found that the oral microbiomes of individuals living in the same household were more similar to each other than to that of individuals in the same city, indicating that geography may stop having a significant influence on the similarity of the oral microbiomes of genetically related individuals after the city level. Shaw et al. also found that individuals who no longer live with their families still had oral microbiomes similar to that of their families, with shared household having a significant effect, and shared genetic variation having no significant effect.
When comparing different methods of sampling for genetic relatedness in the human samples and factoring that into their statistical tests, the authors did find that, when using mapped familial relationships (pedigrees) instead of actual genomic data, genetic relatedness did have a very small, although significant effect on oral microbiome similarity between individuals. Pedigrees, however, can’t reflect the same level of accuracy in measuring genetic relatedness as actual genomic data. For example, a pedigree assumes that, on average, siblings share 50% of their genomic data. Siblings may actually share less than, or in the Ashkenazim case even more than 50% of their genomes due to conservative breeding in the Ashkenazim following the Jewish Diaspora (Atzmon et al. 2010). Thus, due to the robust findings of the tests done on individuals in and between households indicating the effect of household status on the oral microbiome, and the results of the test on individuals no longer living in the same household showing the individuals still had oral microbiomes similar to that of their earlier housemates, the authors ultimately concluded that shared household status had more of an effect on the oral microbiome than genetics.
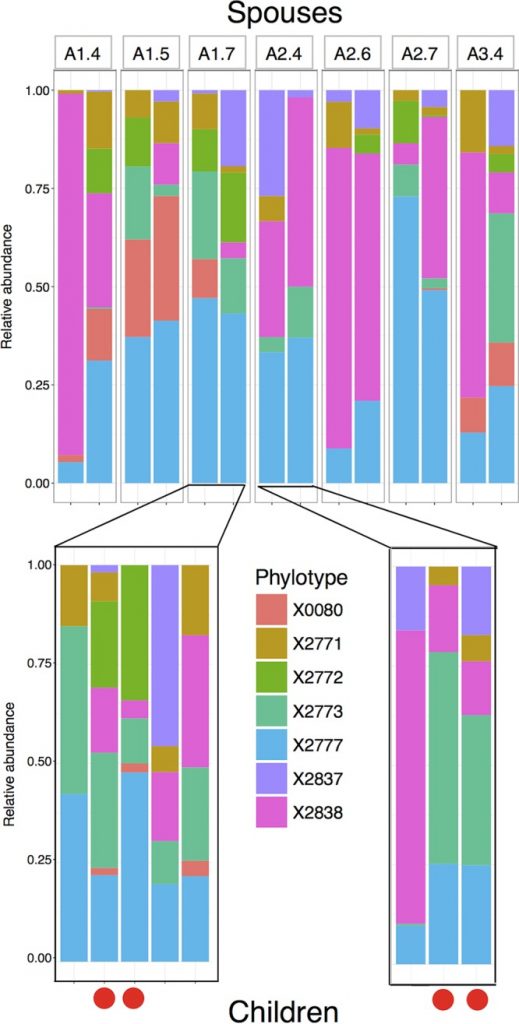
In addition to the previous tests, this study also compared the oral microbiomes of spouses currently living together — individuals genetically dissimilar but living in the same household. While their results weren’t statistically significant, it’s worth noting that, for the most part, the oral microbiomes of spouses were more similar to the spouse they were currently living with rather than an individual living in a different house (Shaw et al. Fig 3). These data indicate that the proximity of spouses living together had more of an effect on the composition of the oral microbiome than the genetics of the spouses. Finally, the study found that individuals who no longer live with their families still had oral microbiomes similar to that of their families, with shared household having a significant effect, and shared genetic variation having no significant effect.
Satisfactory Evidence?
The study by Shaw et al. (2017) used ANOVAs to analyze the effect of multiple variables on the oral microbiome. Some of their results showed findings not consistent, but also not contrary to, their hypothesis. For instance, several of their statistical analyses found age to be a significant factor affecting variation in the oral microbiome. Sequencing plate, the particular batch in a particular run in which their oral microbiome samples were sequenced to detect what organisms were there, also had a significant effect, though both age and sequencing plate had an order of magnitude lower of an effect on the variation than shared household status, indicating it may not have actually had a very large effect on the data. However, for sequencing plate to have any effect, I would question the researchers’ sterile technique. It’s possible contamination between samples on the sequencing plate led it to have a statistically significant effect on their results. This is something to keep in mind for future studies analyzing microbiome composition, as contamination can happen easily in microbial research. In addition, the authors ignored many variables that could affect the oral microbiome, such as underlying diseases and the use of antibiotics. In future studies, I would want to see these variables controlled for in the study by only sampling healthy subjects who haven’t used antibiotics in the recent past.
This study took advantage of a previous study in which an Ashkenazi Jewish family had been genomically analyzed for Crohn’s Disease (Levine et al. 2016). However, what if Shaw et al. had included greater genetic variation in their sampling subjects? Perhaps if genetic variation were greater, there would be more variation available to explain the variation in the oral microbiome, and we would see more pronounced effects of genetics on the microbiome composition.
Along with greater variation, there needs to be an association study between certain spots on the human genome and certain microbes in the oral microbiome before we can definitively say genetic variation plays little to no role in the composition of the oral microbiome. Such studies have already been done on the gut microbiome linking several bacterial species to immune genes in the human genome. (Zhernakova et al. 2016). A replicate of this study, but using fecal samples to analyze the gut microbiome, would be beneficial to our understanding of the role our genetics plays in carving the human microbiome.
This research is the first of its kind to complete an association study between genetics and shared households. It’s not perfect, but it sheds significant light on a previously unknown topic in science. We’re now one step closer to answering the nature versus nurture question with our microbiomes. Do our genetics determine the communities of organisms that live on and in us, or does the environment surrounding us dictate our microbiota? For the oral microbiome, at least, it these findings show how our environment trumps genetics.
Still Curious?
For a general, although technical, overview of microbiome research, watch this talk. If you’re interested in learning more about the oral microbiome, I suggest this talk about the composition of the oral microbiome and how it can affect our health. Keep in mind this talk was done on October 5th, 2015, so the research of this paper isn’t incorporated in the talk. If you’re also interested in the effect the environment can have on your health, mediated by the microbiome of course, I suggest the following articles:
Phillips (2009), which outlines briefly our current knowledge and the direction environmental studies on the human microbiome and our health are going, particularly with the gut microbiome.
US Institute of medicine (2013), which also outlines current knowledge on the environmental effects on the microbiome and how that affects our health. This workshop summary is more technical than the above, but includes a section specifically on the oral microbiome and its unique position in the body.
References
- Atzmon G, Hao L, Pe’ers I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, Ostrer H. 2010. Abraham’s children in the genome era: major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry. The American Journal of Human Genetics: 86;6; 850-859. doi:10.1016/j.ajhg.2010.04.015
- Costa, D. Marta et al. 2013. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nature Communications: 4; 2543. doi:10.1038/ncomms3543
- Institute of Medicine (US) Food Forum. Interaction Between the Microbiome and Health and Environment The Human Microbiome, Diet, and Health. 2013. Workshop Summary; Washington (DC): National Academies Press (US); Published February 22-23, 2013.
- Levine AP, Pontikos N, Schiff ER, Jostins L, Speed D, NIDDK Inflammatory Bowel Disease Genetics Consortium, Lovat LB, Barrett JC, Grasberger H, Plagnol V, Segal AW. 2016. Genetic complexity of Crohn’s disease in two large Ashkenazi Jewish families. Gastroenterology 151:698—709. doi:10.1053/j.gastro.2016.06.040
- Phillips, M. L. 2009. Gut Reaction: Environmental Effects on the Human Microbiota. Environmental Health Perspectives, 117(5), A198—A205. doi:10.1289/ehp.117-a198
- Proctor, L. 2016. Human Microbiome Research: Where We Are and Where We Might Go. Workshop recording. Environment and health: what’s the Human Microbiome got to do with it? The National Academies of Sciences, Engineering, and Medicine. Published January 21, 2016.
- Shaw L, Ribeiro ALR, Levine AP, Pontikos N, Balloux F, Segal AW, Roberts AP, Smith AM. 2017. The human salivary microbiome is shaped by shared environment rather than genetics: evidence from a large family of closely related individuals. doi:10.1128/mBio.01237-17
- Winick, R. October 5, 2015. The oral microbiome: friend or foe? Functional Forum. Published October 28, 2015.
- Zhernakova A, Kurilshikov et al. 2016. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352:565—569. doi:10.1126/science.aad3369
