
Background:
About 70% of the planet is covered in water with about 60% of your body being made up of water. Lakes, rivers, and reservoirs contain some of the most diverse and abundant communities of bacteria according to a 16S rRNA gene study by Tamames et al. (2010). Our own microbiota interact daily with these different communities as we drink, bathe, and excrete bacteria. One of the more pressing issues in today’s modern medicine is antibiotic resistant bacteria that are becoming increasingly hard to treat. Antibiotic resistance is caused by evolution of bacteria, which is the bacteria gaining antibiotic genes that increase its fitness. In the presences of antibiotics, bacteria that are lacking a resistance mutation die, whereas bacteria with immunity to antibiotics have a higher survival rate. These antibiotic resistant bacteria then pass on that resistance to antibiotics to the next generation, as well as potentially other bacteria through lateral gene transfer, which is the transfer of genetic material between a parent and daughter bacteria.
Central Question:
How does bacteria share antibiotic resistance across the human created water cycle? What is the impact on human health?
Evidence:
In this article the authors, Ivone Vaz-Moreira et al.(2014) focused on what role bacteria plays in the sharing of antibiotic resistance across the human created water cycle and its impact on human health. They looked at how we share and host different bacteria with water sources, and the potential pathways bacteria have when crossing from one source to another. The authors also looked to find out which specific bacteria are intermediates, donors, or receptors when passing genes of antibiotic resistance in the pathways between humans and the water cycle.
There have been multiple studies conducted that have shown that, while there are harmless bacteria in our water ecosystems, there is also bacteria present that is resistant to antibiotics, especially in our wastewater treatment plants (Manania et al., 2011; Rizzo et al., 2013). Ivone Vaz-Moreira et al. (2014) suggests lateral gene transfer (LGT) occurs among microbes in our own drinking and waste water, which increases antibiotic resistance in bacteria. According to research by Gillings et al. (2013), the antibiotics that we use to treat bacterial infections have a longer half-life than the time spent in our body. As a result, many antibiotics end up in our water systems as a waste product. Gillings et al. (2013) actually considers antibiotics as pollutants, because they interact with the bacteria in those water systems. This increases the recombination or shifting around the genes, which increases mutations and LGT to amplify resistant bacteria. There are three different responses that bacteria have been shown to have towards antibiotics either through silent, acquired, or intrinsic resistance (Alvarez-Ortega et al., 2011). Intrinsic resistance is a preexisting tolerance that gives the bacteria mechanisms to resist the affects of antibiotics, these genes however cannot be transferred through lateral gene transfer easily. Acquired resistance is when the bacteria is able to adapt through recombination/mutations to an antibiotic agent it was previously vulnerable to, as well as LGT from other antibiotic resistant bacteria through the form of plasmid vectors. In silent resistance, the bacteria have genes that aren’t resistant to antibiotics in the host bacteria but, when transferred to other bacteria, they offer resistance. Many of the genes that are antibiotic resistant are in the form of integrons or plasmids that can be integrated and utilized quickly in response to antibiotics.
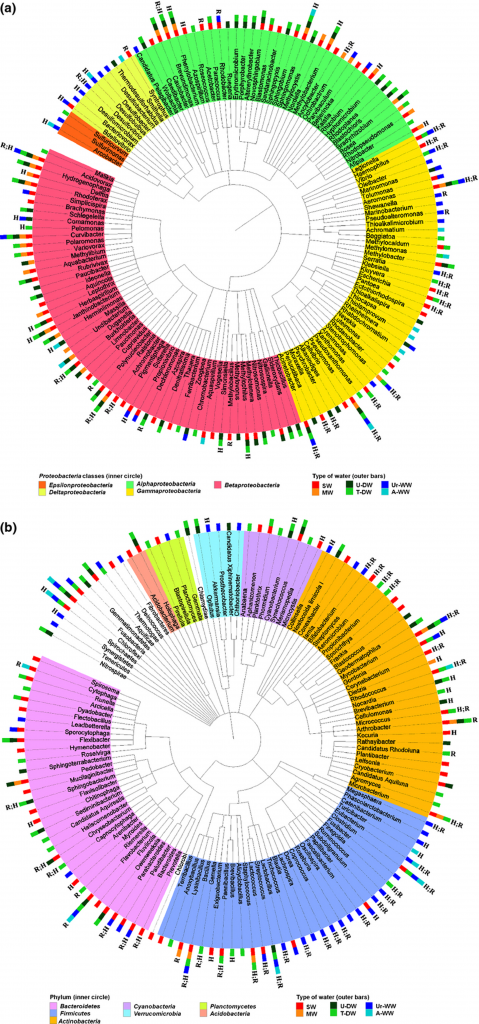
Previous studies have also found that waste treatment plants had multitudes of antibiotic resistant bacteria, and that the treatments are unable to completely disinfect and remove them (Manaia et al., 2011). They have found that this is the primary source of human caused antibiotic resistance in bacteria. Even though this is the cause of multiple resistant bacteria, such as E. Coli and Enterococcus, it is not the only source of antibiotic resistant bacteria. According to Massa et al. (1995), they found multiple antibiotic resistant bacteria in the commercial spring and mineral waters that are sold to the public that come from natural water sources. Mineral water and Spring water are one of the more popular drinks that humans consume, but the problem is that there is no way to remove or disinfect the bacteria that is present due to the regulations by the European Commision, and the water has microbiota that, when consumed, interacts with the human microbiota. There have also been several bacteria that have been shown to show a high risk of developing antibiotic resistance that are in drinking water and the human microbiome such as Clostridium, Acinetobacter, and Aeromonas (Zhang et al., 2009).
In other studies, they found that there are already numerous antibiotic bacteria that are already present in the human gut resistome (collection of all antibiotic genes in the human gut) that are different from already sequenced bacteria that we’ve known of (Sommer et al., 2009). The human microbiome has been suggested by Sommer et al. (2009) as being the connection between environmental resistomes(a collection of antibiotic resistance genes) and pathogens present in humans with the human microbiome containing multiple antibiotic resistance bacteria. This would make sense with humans consuming water contaminated with antibiotic resistant bacteria and interactions with the human gut resistome could show a transfer of antibiotic genes. However not all bacteria strains are able to thrive in both water environments and the human body, which could be an explanation for only a fraction of the human microbiota being similar to bacteria in water sources.
Questions:
1. There is difficulty culturing bacteria, which could exclude many types of bacteria that may be contributing to the pathway of antibiotic resistance. What other bacteria could be playing a role in antibiotic resistance in the water systems?
2. There has also been evidence in studies, such as Zhang et al. (2009), showing that microbes in our own microbiome are present in natural water habitats. What could their role be in transmitting antibiotic resistance genes?
Further Reading:
(Allen et al., 2009)– In this study they look at Alaskan soil in remote locations without human influences and found that beta-lactamases develop resistance genes that could potentially transfer to E. Coli, which could be a potential pathogen.
(Riesenfeld et al., 2004)– In this study researchers looked at the bacteria that could have antibiotic resistance that could have been missed when soil samples are cultured in the lab with PCR. They were able to isolate DNA sequences from the soil directly and then clone the sequences of genes of antibiotic resistance and use culture-independent methods to find there are a greater diversity of bacteria with antibiotic resistance than what has been previously found.
(Zhang et al., 2009)-In this study they look at different research on the emergence of antibiotic resistance genes in water sources such as hospital waste water and animal farms waste-water. They look at the different ways to isolate antibiotic resistance genes with specific and multiplex PCR.
Resources:
- Ivone Vaz-Moreira, Olga C. Nunes, Célia M. Manaia; Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome, FEMS Microbiology Reviews, Volume 38, Issue 4, 1 July 2014, Pages 761—778, DOI:10.1111/1574-6976.12062
- Gillings, M. R. (2013). Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4. DOI: 10.3389/fmicb.2013.00004
- Massa S, Petruccioli M, Fanelli M & Gori L (1995) Drug resistant bacteria in non carbonated mineral waters. Microbiol Res 150: 403—408. DOI: 10.1016/S0944-5013(11)80022-4
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I & Fatta-Kassinos D (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447: 345—360. DOI: 10.1016/j.scitotenv.2013.01.032
- Shi P, Jia S, Zhang XX, Zhang T, Cheng S & Li A (2013) Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res 47: 111—120. DOI: 10.1021/acs.est.5b03521
- Sommer MO, Dantas G & Church GM (2009) Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325: 1128—1131. DOI: 10.1126/science.1176950
- Zhang XX, Zhang T & Fang HH (2009) Antibiotic resistance genes in water environment. Appl Microbiol Biotechnology 82: 397—414. DOI: 10.1007/s00253-008-1829-z.
- European Comission (2009) DIRECTIVE 2009/54/EC — on the Exploitation and Marketing of Natural Mineral Waters (Recast). European Comission, Official Journal L 164, 26-6-2009, 0045-0058. Brussels. https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32009L0054
- Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW & Martınez JL (2011) The intrinsic resistome of Pseudomonas aeruginosa to b-lactams. Virulence 2: 144—146. DOI: 10.1128/AAC.01583-12
- Allen HK, Moe LA, Rodbumrer J, Gaarder A & Handelsman J (2009) Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3: 243— 251. DOI: 10.1038/ismej.2008.86
- Riesenfeld CS, Goodman RM & Handelsman J (2004) Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol 6: 981—989. DOI: 10.1111/j.1462-2920.2004.00664.x
-
- Manaia CM Vaz-Moreira I Nunes OC (2011) Antibiotic resistance in waste water and surface water and human health implications. The Handbook of Environmental Chemistry , Chapter 6, Vol. 20 (Barceló D, ed), pp. 173—212. Springer-Verlag, Berlin, Heidelberg. DOI: 10.1007/698_2011_118
- Manaia CM Vaz-Moreira I Nunes OC (2011) Antibiotic resistance in waste water and surface water and human health implications. The Handbook of Environmental Chemistry , Chapter 6, Vol. 20 (Barceló D, ed), pp. 173—212. Springer-Verlag, Berlin, Heidelberg. DOI: 10.1007/698_2011_118
