Background
The microbiome of the gut specifically plays a role in chronic metabolic disease, one of which is Chronic Kidney Disease (CKD) (Ren et al. 2020) which this blog post specifically covers. CKD is a chronic condition because the damage to kidneys occurs over a long time period, inhibiting their ability to filter blood and toxins. As the damage occurs over time there is increasing risk that patients will end up on dialysis or need transplantation. The sooner CKD can be diagnosed the higher chance a patient has to protect their kidneys (“What is chronic Kidney disease?” 2017). CKD is an important disease to discuss since approximately 13.4% of the global population suffers from it and around 30 million people in the U.S. (Ren et al. 2020). Sufferers have a greatly increased risk of morbidity and mortality, as well as suffering from the significant healthcare costs that arise with this disease. For most patients CKD is not diagnosed until it is in a very progressed stage because the clinical symptoms are normally nonexistent in the early stages, meaning that most end up in end-stage renal failure (ESRF) which requires dialysis, transplantation, or other costly and long-term medical procedures. In 2012, Viziri et al. demonstrated the relationship between the gut microbiome and CKD. Gut derived uremic toxins, created by enzymes that the microbiota of the gut harbor are a factor in the progression of CKD and in previous studies it has been observed that as renal function decreases there is an increase in these toxins. Even though we know the connections with advanced stages of the disease, there is little research that has been done to potentially use the microbiome to diagnose early stage CKD, which leads to the researchers main question.
Central Question
What are some diagnostic markers in the gut microbiota that could signify CKD? There are little clinical signs of CKD usually until it is too late, so for many patients they end up in ESRF where their prognosis is discouraging. identifying contributing microbial taxa and utilizing that discovery to create a diagnostic tool could lead to much earlier detection and a better prognosis for patients.
Evidence
This study was performed in two regions of China (Ren et al. 2020). 489 total fecal samples were collected and sequenced using DNA sequencing methods: 216 of the samples were from patients with CKD and 273 were from healthy individuals (HC). Three different groups were used in the study: a discovery cohort (used to construct a CKD classifier); a validation cohort (used to verify the efficacy of the classifier); and an independent diagnosis group of a number of other CKD patients to further evaluate efficacy of the classifier that was developed. The discovery cohort was further divided into three groups depending on the stage of CKD. In the discovery cohort different diversity indices were used to indicate differences between the healthy and diseased samples, in regards to their microbiota composition. The results showed the gut microbial diversity of the CKD samples were greatly decreased compared to the HC ones, as well as the observed taxonomic units (OTUs) which can be described as the number of closely related bacterial groups present. When the compositions were compared at all different levels (phyla, genera, class, order, family) there were significant differences in the diversity of the microbiota between the healthy and CKD samples as shown by Figure 1. below.
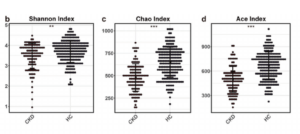
A model was developed to choose specific markers that, if detected in a sample could signify that a patient had CKD. Using this model the researchers identified five OTUs as optimal markers of CKD. The model was used to generate values that signified the probability of having CKD called the probability of disease (POD) and the POD values of the CKD samples were significantly higher than those of HC samples, suggesting that these markers were identifying CKD patients. This diagnostic tool was used in the validation and then independent groups of samples where it again showed greatly increased POD values for the CKD samples, verifying the results. There was not a significant difference in microbial diversity between the three discovery groups but some differences seen such as an increase in Akkermansia is thought to play a role in the progression of CKD. Further investigation into the specific changes as CKD progresses could lead to future specific treatments. Overall the researchers noted that as CKD develops and progresses it is noted that the microbial community changes significantly. They matched some taxa which were prevalent in CKD patients to enzymes whose functions, among others, include production of uretic toxins that can be used as markers for CKD progression. It is thought that an overgrowth of these taxa could facilitate CKD progression by affecting the synthesis or uremic toxin molecules. Being able to identify these taxa in the early stages could prove to be life changing for many patients in the future.
My Questions
This study was performed in China so I would be interested to see if the markers would be the same in other countries and areas of the world. Would they be similar? Would they be Different? Could the same tool be used? Or would there need to be different tools depending on ethnicity and location of the patient? Fecal studies using stool samples have been used in numerous other studies so I believe it would be possible to potentially match the markers found in this study to the data from others. Furthermore I believe over time extensive databases can be built as microbial testing continues and data begins to accumulate that will make it easy for researchers to reference their findings with other data.
Another question I identified was regarding the future studies concerning the problematic taxa. It could be beneficial to work towards further identifying and developing ways to eliminate them within the microbiota upon the onset of CKD. If feats such as this could become possible it would be a huge progression in medicine and treatment of disease.
Further reading
There are many website sources that provide good information on the basics of chronic kidney disease; the signs, symptoms, treatments etc. Below are a few good papers that further highlight and review the research done on the relationship between the gut microbiome and these chronic diseases.
Wing, M. R., Patel, S. S., Ramezani, A., & Raj, D. S. (2015). Gut microbiome in chronic kidney disease. Experimental Physiology, 101(4), 471-477. doi:10.1113/EP085283
Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res. 2017 Jan;179:24-37. doi: 10.1016/j.trsl.2016.04.007.
If you just want to learn more about what CKD is and the symptoms, causes and treatments here are a few websites that have good factual information.
Chronic Kidney disease information from Mayo Clinic
Some facts on Chronic Kidney disease from the National Kidney Foundation
References
- Sordi, V., MG. Rooks, W., DY. Li, W., B. Afsar, N., R. Liu, J., H. Wu, E., . . . D. Viramontes-Horner, F. (1970, January 01). Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease. Retrieved October 12, 2020. doi: 10.1186/s12967-018-1756-4
- Ren, Z., Fan, Y., Li, A., Shen, Q., Wu, J., Ren, L., . . . Li, L. (2020). Alterations of the Human GutMicrobiome in Chronic Kidney Disease. Advanced Science, 2001936. doi: 10.1002/advs.202001936
- Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen TH, Andersen GL. (2012, February). Chronic kidney disease alters intestinal microbial flora. Kidney Int. doi: 10.1038/ki.2012.345.
- What Is Chronic Kidney Disease? (2017, June 01). Retrieved September 25, 2020.
