Background:
What is IBS? Irritable Bowel Syndrome is a functional bowel disorder. Functional GI (gastrointestinal) Disorders are defined as being related to how your gut and brain work together: this can make your intestines more sensitive, which can change how the muscles associated with your gut contract. IBS is most often associated with abdominal pain and cramping, changes in bowel movement and excess gas. Though IBS is non-fatal there are significant impacts on the quality of life of those affected, leading to mobility issues, chronic pain, and increases in anxiety and depression. While IBS is often a diagnosis of exclusion and the exact cause is not known, some risk factors of IBS are family history; if you are young (under the age of 50), are female, or have mental health issues, like anxiety or depression (1). IBS has also been known as a spastic colon, spastic colitis, and mucous colitis.
IBS can have symptoms similar to Inflammatory bowel disease (the most common of which are Crohn’s Disease and Ulcerative Colitis), but they are not the same. The theorized cause of IBS ranges from muscle contraction in the intestines that are different from normal. Stronger contraction can last longer, cause gas, bloating, and diarrhea; whereas weaker contraction can cause slowed food passage and constipation (both of which are types of IBS) (2). Serotonin is a way your brain talks to your intestines (though that is not all it does) can impact the movement of the muscles around the gut, leading to the same problems. Another theorized cause is a disorder known as SIBO (Small Intestine Bacterial Overgrowth). Diagnosis of IBS often occurs after colonoscopies, endoscopies, and x-rays or CT scans of the abdominal area.
In 80% of cases food has been identified as a trigger, and the FODMAP diet has been reported to provide substantial relief in 50-70% of cases (3). So, what is the FODMAP diet? FODMAP is an acronym for “Fermentable Oglio-, Di- Mono-saccharides, and Polyols.” These are short chain carbohydrates that the small intestine doesn’t like to absorb, so they absorb water and are fermented by your gut bacteria. However, some people have more difficulty digesting them than others, and this can contribute to IBS symptoms. The question “how does that impact the gut microbiome?” remains. What we eat impacts what the microbes in our intestines eat, and that impacts who lives there (4).
Previous studies have shown differences in the microbiomes of people with IBS and those without (5). These differences can be characterized by both an overabundance and a much lower diversity of microbes in the intestines (6).
Because IBS is a functional bowel disorder, the interaction between the intestines and the brain are abnormal. This gut-brain axis (a term that refers to how the brain and the intestines interact) has been hypothesized to be a microbe-gut-brain axis, which includes the central nervous system (CNS), the neuroendocrine and neuroimmune systems, both the sympathetic and parasympathetic limbs of the autonomic nervous system, the enteric nervous system (ENS) and, finally, the gut microbiome. In other words, the microbiota can affect nerves in the intestines, which talk to the brain, and the brain talks to the intestines, which impacts the environment the microbes live in. This relationship has led to the belief that our gut microbiome can, and does, interact with nearly all parts of our biology. It is implied that the gut microbiome can even control their host’s endocrine systems (our hormones) (7).
Central question:
What are the differences between the microbial structures of those with IBS and those without?
Evidence:
The duodenum is the beginning of the small intestine, directly following the stomach.
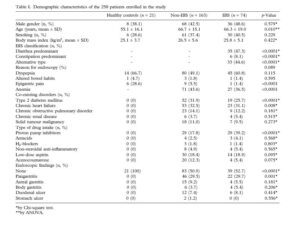
Giamarellos-Bourboulis et al (2015) sampled a total of 21 healthy patients, 163 non-healthy patients, and 74 IBS patients. Duodenal aspirates, which are biopsies of the mucosa in the intestines, were taken during colonoscopy procedures.
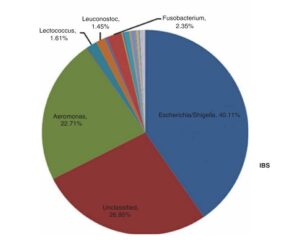
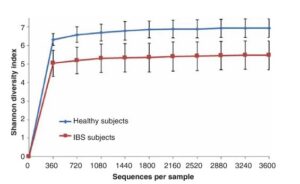
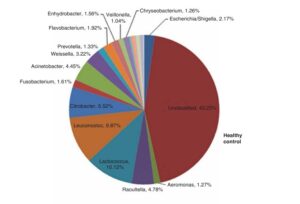
The DNA of the samples were extracted and then sequenced in two different ways. The first method is 16S sequencing, which can tell researchers “who is there.” Giamarellos-Bourboulis et al found an increase in Escherichia and Aeromonas species, and decreases in Acinetobacter, Citrobacter, Leuconostoc, and Lactococcus species when compared to healthy controls. All of these are normal members in the gastro-intestinal tract, it is not the presence, but the proportion of these species that were of interest. Giamarellos-Bourboulis et al calculated a Shannon Diversity Index, a measure that looks at how many different species are in a data set, for both IBSers and the healthy controls and found statistically significant differences where IBSers had far less diversity. Conversely, the intestines of those with IBS were found to have more microbes overall. Finally, different types of IBS could be associated with the overabundance of different types of microbes. IBS-C (IBS with constipation) was characterized by overabundance of Escherichia/Shigella species, while IBS-D (IBS with diarrhea) was associated with under-representation of Escherichia/Shigella species and over representation of Aeromonas species, but these were not significant statistically.
The second method of DNA sampling is qPCR (quantitative PCR), which measures the differing amounts of DNA in specific samples. With this method the authors were looking for specific differences between those with IBS and those without. They found specific increases in Escherichia. coli and K. pneumoniae in IBSers. This test confirmed small intestine bacterial overgrowth (SIBO) correlation with IBS.
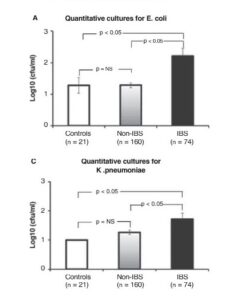
This study was the first to use modern DNA sequencing techniques to look at the small intestine in regards to IBS. Giamarellos-Bourboulis et al found that IBS can be characterized by an overabundance of microbes that are less diverse than healthy intestines, as can be seen in the graphs below.
My questions:
1. The question of “what causes IBS” remains unanswered, could the over representation of E. coli be related to it?
Finding the cause of IBS would greatly impact how we diagnose it and how we treat it. E. coli is a normal part of the gut microbiome, it can even help us synthesize vitamin K, but it can cause food poisoning. IBS, in 3 to 36% of cases is preceded by inflammation of the bowel, this (along with other findings) could point to pathogenesis of E. coli as the cause of IBS. (DOI: 10.1093/femsle/fny249)
2. Why is SIBO (small intestine bacterial overgrowth) related to both IBS and Diabetes?
This was a linkage in the paper, but was not expanded upon. It was referenced in relation to a table that was not included with the paper.
3. How are IBS (Irritable Bowel Syndrome) and IBD (Inflammatory Bowel Disease) related?
IBD is an umbrella term for chronic inflammatory disorders of the digestive tract. Ulcerative Colitis and Crohn’s Disease are both types of IBD. There are several similarities between IBS and IBD that bring into question the differences between the two. For example, immune responses, microbial imbalance in the gut are reminiscent of IBS. Additionally, when severe IBD is medicated symptoms similar to IBS can be observed.
Further reading:
IBS and IBD — separate entities or on a spectrum?
By Robin Spiller and Giles Major in Nature Reviews Gastroenterology and Hepatology, 26 September 2016. DOI: 10.1038/nrgastro.2016.141
Are Irritable Bowel Syndrome and Inflammatory Bowel Disease two separate diagnoses, or are they the same? The treatments of IBS, dietary regulation generally, could help those who suffer from IBD.
Damage of the Mucus Layer: The Possible Shared Critical Common Cause for Both Inflammatory Bowel Disease (IBD) and Irritable Bowel Syndrome (IBS)
By Xiaofa Qin in Inflammatory Bowel Diseases, Volume 23, Issue 2, 1 February 2017. DOI: 10.1097/MIB.0000000000001010
In recent scholarship there has been a linking of Inflammatory bowel diseases and Irritable Bowel Syndrome. Traditional medicinal thought these two are thought to be separate, based on several diagnostic features, but there has been recent scholarship that may tie the two diagnoses as opposite ends of the same spectrum. The author argues that these two have a shared cause.
References:
- “Irritable Bowel Syndrome.” Mayo Clinic, Mayo Foundation for Medical Education and Research, 15 Oct. 2020, www.mayoclinic.org/diseases-conditions/irritable-bowel-syndrome/symptoms-causes/syc-20360016.
- Herndon, Jaime. “IBS: Symptoms, Causes, Diagnosis, Triggers, and Treatment.” Healthline, Healthline Media, 8 Mar. 2019, www.healthline.com/health/irritable-bowel-syndrome.
- Selvaraj, Shabnam Mohajir, et al. “Role of Low FODMAP Diet and Probiotics on Gut Microbiome in Irritable Bowel Syndrome (IBS).” Progress In Microbes & Molecular Biology, vol. 3, no. 1, 2020, doi: 10.36877/pmmb.a0000069.
- Brody, Herb. “The Gut Microbiome.” Nature News, Nature Publishing Group, 29 Jan. 2020, doi: 10.1038/d41586-020-00194-2
-
Intestinal microbiota in functional bowel disorders: a Rome foundation report
- Reddel, Sofia, et al. “The Impact of Low-FODMAPs, Gluten-Free, and Ketogenic Diets on Gut Microbiota Modulation in Pathological Conditions .” Nutrients, vol. 11, no. 2, ser. 373, 4 Feb. 2019. 373, doi: 10.3390/nu11020373.
- Kennedy, Paul J. “Irritable Bowel Syndrome: A Microbiome-Gut-Brain Axis Disorder?” World Journal of Gastroenterology, vol. 20, no. 39, 21 Oct. 2014, p. 14105., doi: doi.org/10.3748/wjg.v20.i39.14105.
- Chang, Lin, and David Geffen. “Irritable Bowel Syndrome (IBS).” National Institute of Diabetes and Digestive and Kidney Diseases, U.S. Department of Health and Human Services, www.niddk.nih.gov/health-information/digestive-diseases/irritable-bowel-syndrome.
- Dogan, Belgin, et al. “Evaluation of Escherichia Coli Pathotypes Associated with Irritable Bowel Syndrome.” FEMS Microbiology Letters, 8 Oct. 2018, doi: 10.1093/femsle/fny249.
- Eisenstein, Michael. “Microbiome: Bacterial Broadband.” Nature, vol. 533, no. 7603, 18 May 2016, doi: doi.org/10.1038/533s104a.
- Giamarellos-Bourboulis, Evangelos, et al. “Molecular Assessment of Differences in the Duodenal Microbiome in Subjects with Irritable Bowel Syndrome.” Scandinavian Journal of Gastroenterology, vol. 50, no. 9, 2015, pp. 1076–1087., doi: 10.3109/00365521.2015.1027261.
- Sikander, Arbab, et al. “Role of Serotonin in Gastrointestinal Motility and Irritable Bowel Syndrome.” Clinica Chimica Acta, vol. 403, no. 1-2, May 2009, pp. 47–55., doi: 10.1016/j.cca.2009.01.028