Background
Colorectal Cancer (CRC) makes up a large proportion of cancer in the United States. CRC occurs in two locations, the colon and rectum. It is the 3rd most diagnosed type of cancer in the U.S., and in 2021 it is estimated there will be just over 100,000 cases of colon cancer and around 45,000 cases of rectal cancer leading to more than 50,000 deaths (American Cancer Society, 2021).
Colorectal cancer causes the third most cancer-related deaths among men and women separately, and if you combine into one population it causes the second most. The actual death rate of CRC has decreased in the last couple of decades due to increase in effectiveness and quantity of screenings (e.g. colonoscopy) which look for signs of CRC, including polyps (small clump of cells that can be cancerous) located in the rectum/colon (Stewart and Carter-Templeton, 2017). Even with the ability to screen and look for physical changes in the colon/rectum, many people still die from CRC and it is a major problem facing the field of oncology and medicine in general.
In recent years, more and more research is investigating the impact of the human microbiome on human health and disease (Cong and Zhang, 2018). Specifically, a large amount of research focuses on better understanding the makeup of the gut microbiome, as well as its function. These functions include digestion processes as well as possible immune function.
Researchers are looking at how the gut microbiome influences certain diseases as well as the immune system and finding that the gut microbiome does impact human health more than just as a digestion influencer but in the immune response as well (Clemente et al., 2018). This review provides a detailed list of the many diseases possibly impacted by the composition of the gut microbiome, as well as certain projects and projects to come that look at the correlations between the gut microbiome and human diseases.
This idea that the gut microbiome can influence the immune response of someone supports the idea that it can possibly help determine whether a certain disease is present, and possible risk factors for a disease. If a person has a biomarker, a high concentration of a specific microbe associated with a certain disease, this would lead to the conclusion that the patient probably has the disease. Also, if a patient is lacking certain microbes associated with fighting diseases or has high concentrations of a disease-causing microbe then these would be risk factors of contracting/developing a disease.
Central Question
Are there microbiome biomarkers that will indicate possible CRC cases? While conducting a meta-analysis using the data from multiple studies located around the world, Wirbel et al (2019) hypothesized that certain specific species would be present in all CRC patients that could serve as signatures for CRC, and possibly lead to a diagnosis. They also wondered what types of genes would be present in the CRC specific microbes and what different types of functions the genes would have.
Evidence
Wirbel et al (2019) wanted to compare the gut microbiomes of CRC patients with control non-CRC positive patients to determine if there were differences in micobe abundance. To do this, they used eight unique geographical shotgun metagenomic studies (shotgun metagenomic sequencing is a process that allows researchers to look at all the genes in every organism in a sample in order to look at bacterial diversity and abundance and provides a way to study unculturable microbes) and identified 29 species that were enriched in CRC metagenomes (collection of genetic material from a community of mixed organisms in a specific location, like the colon/rectum).
The studies were located in 7 different countries around the world including France, Austria, China, United States, Germany, Italy, and Japan. The meta-analysis used four published studies that included shotgun metagenomics to compare healthy and CRC patients. The fifth study, located in Germany, was a new study that the meta-analysis created to generate new fecal metagenomic data for CRC and non-CRC patients in Germany.
The studies included different sampling procedures, storage processes, and DNA extraction protocols, but all of them included fecal metagenomic data from CRC patients and non-CRC patients to create a larger sample size (386 CRC patients and 392 tumor free controls) with samples from around the world.
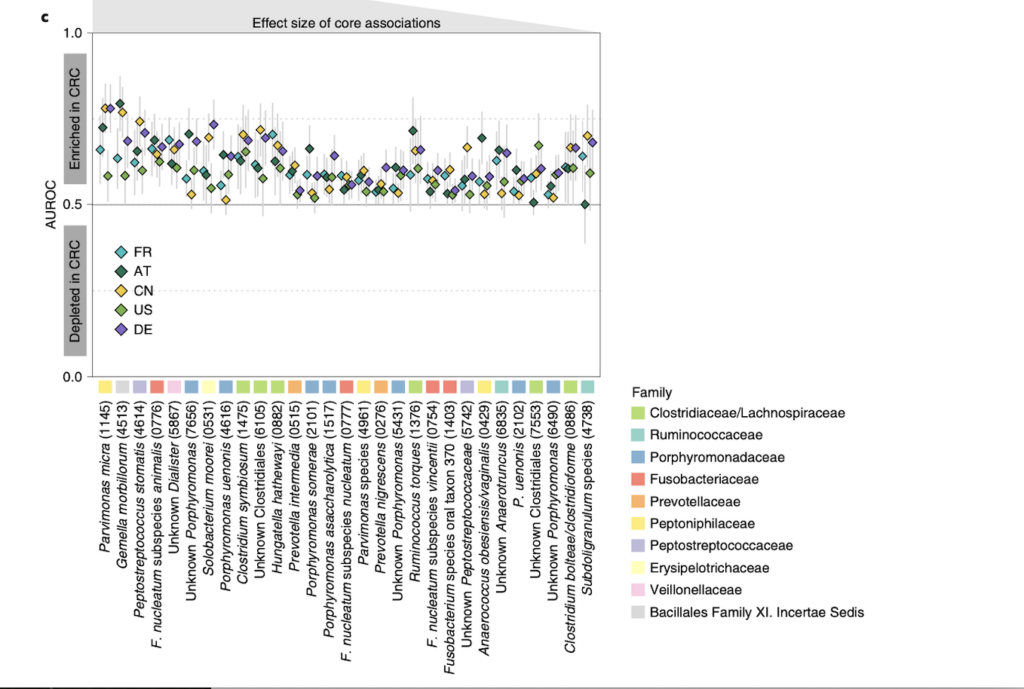
Analyzing the patients microbiomes from each location, the researchers were able to identify a core set of 29 species of bacteria that were significantly enriched or more abundant in CRC patients compared to healthy ones. A list of the 29 species is located in Fig. 1 (located bottom of evidence section), and it conveys how the 29 species were significantly enriched in CRC patient gut microbiomes. This supports their final conclusion that these 29 species can be used as possible markers for CRC due to the overall presence of these microbes in CRC patients.
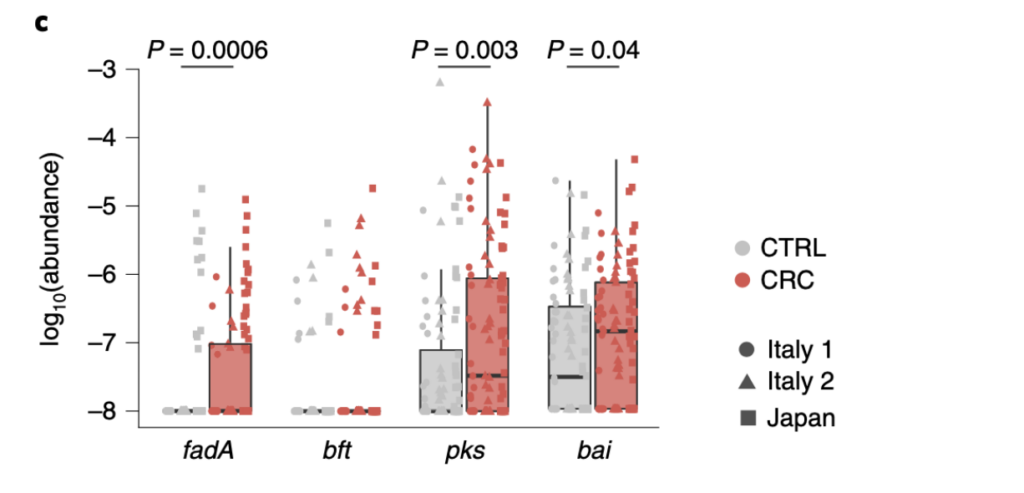
Another idea that was discussed in the paper is that certain genes might be present in higher levels in the gut microbiome of CRC patients. They did find 3 genes to be enriched in CRC patients. The genes are fadA, pks, and bai. The support for these genes being present in higher levels than control patients is located in Fig. 2 (located at the bottom of the evidence section).
The sequence data from three locations (Italy 1, Italy 2, and Japan) showed significant enrichment of the fadA, pks, and bai in CRC patients. These genes are associated with carcinogenic effects. For example, the study confirmed highly significant enrichments of the colibactin-producing pks gene cluster as well as FadA in CRC metagenomes supporting the clinical relevance of these genes and adds support for the experimental evidence for their carcinogenic effect. Another example, is the paper discusses 7alpha-dehydroxylation is encoded in the bai operon. This pathway is associated with liver cancer and now is hypothesized to also promote CRC.
Questions
One area I would like to look at would be the actual microbiome located in/on tumors. While reading other papers, I discovered studies looking at the possible implications of actual tumor microbiomes and their positive and negative impact on malignant tumor growth/development (McAllister et al., 2019). For example, a study could look at dissecting tumors/polyps located in the colon/rectum of patients and look at the microbiome of these samples. Perhaps there will be a link between certain microbes and the aggressiveness of certain cancers in specific patients. Not every case of CRC is the same, some are more aggressive than others, and I am curious if the tumor microbiome or human microbiome in general has an impact on this.
Another area I would like to look into would be possible implications of a study like this looking at other cancers besides CRC. The topic of correlation between the gut microbiome and human health has been heavily studied not just for CRC but for digestion, obesity, IBD, and so many other diseases. I would like to research possible new locations (other high risk cancer locations in the body) and see if there are microbe signatures. Much of cancer research goes into looking at how to treat and limit cancer development, but maybe there is a larger connection with our microbiome and cancer. The study above supports the idea that the gut microbiome has a connection with CRC, I think it would be worth looking into the microbiomes of other areas for possible connections.
For this study specifically, they briefly talk about future studies involving their research and the one area I thought was interesting was looking at possible virome carcinogens. Viruses are an important microbe in our microbiome and some are known to have cancer causing effects. For example, the Human Papillomavirus (HPV) is known to cause cancer, specifically cervical cancer. I think another area to look into is the virome, the collection of viruses found in certain areas like the gut, and see whether or not there are markers that can be found to possibly diagnose/prevent CRC and other types of cancer.
Further Reading
This paper is a review of papers about how the microbiome is connected with cancer and cancer therapy.
- Helmink, B. A., Khan, M. W., Hermann, A., Gopalakrishnan, V., & Wargo, J. A. (2019). The microbiome, cancer, and cancer therapy. Nature medicine, 25(3), 377-388. DOI:10.1038/s41591-019-0377-7
This paper discusses possible implications of the microbiome on breast cancer.
- Plaza-Díaz, J., Álvarez-Mercado, A. I., Ruiz-Marín, C. M., Reina-Pérez, I., Pérez-Alonso, A. J., Sánchez-Andujar, M. B., … & Fontana, L. (2019). Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC cancer, 19(1), 1-9. DOI: 10.1186/s12885-019-5660-y
This paper is similar to the meta-analysis looking at potential microbe biomarkers for CRC.
- Yao, Q., Tang, M., Zeng, L., Chu, Z., Sheng, H., Zhang, Y., … & Ye, M. (2021). Potential of fecal microbiota for detection and postoperative surveillance of colorectal cancer. BMC microbiology, 21(1), 1-11. DOI: 10.1186/s12866-021-02182-6
References
- Clemente, J. C., Manasson, J., & Scher, J. U. (2018). The role of the gut microbiome in systemic inflammatory disease. Bmj, 360. DOI: 10.1136/bmj.j5145
- Colorectal cancer information: Understanding colorectal cancer. American Cancer Society.(2021, January 12).
- Cong, J., & Zhang, X. (2018). How human microbiome talks to health and disease. European Journal of Clinical Microbiology & Infectious Diseases, 37(9), 1595-1601. DOI:10.1007/s10096-018-3263-1
- McAllister, F., Khan, M. A. W., Helmink, B., & Wargo, J. A. (2019). The tumor microbiome in pancreatic cancer: bacteria and beyond. Cancer cell, 36(6), 577-579. DOI:10.1016/j.ccell.2019.11.004
- Stewart, R., & Carter-Templeton, H. (2017). A study comparing colorectal cancer screening techniques. Gastroenterology Nursing, 40(2), 121-127. DOI:10.1097/SGA.0000000000000207
- Wirbel, J., Pyl, P. T., Kartal, E., Zych, K., Kashani, A., Milanese, A., … & Zeller, G. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature medicine, 25(4), 679-689. DOI: 10.1038/s41591-019-0406-6
