Background
Microbes are everywhere. They live in our backyards, on our pets, in our homes and even on and inside of us! Microbes are the bacteria, fungi and viruses that exist in a particular environment, including the human body, soil, plants and the kitchen counter (Marchesi & Ravel, 2015). Our first experience with microbes is usually at home, where we are taught to wash our hands to prevent us from getting sick. “You can’t eat without washing,’ your aunt might say, “there are germs all over your hands!’ Your aunt is right. For every human cell in our body, there is a microbe cell to match it (Sender et al., 2016). However not all of these microbes are harmful.
Many microbes are known to cause infection and disease, but there are many microbes that help us maintain healthy bodies (Human Microbiome Consortium, 2010), enhance digestion (Tilg & Kaser, 2011), synthesize vitamins (Jincai et al., 2009) and prevent harmful pathogens from colonizing us (Human Microbiome Project). Microbes tend to be more associated with disease and sickness, but are actually responsible for keeping us healthy (Human Microbiome Consortium, 2010). What if many common diseases aren’t actually by a specific pathogen, but rather imbalances of the microbial communities living in our bodies? That’s why the Human Microbiome Project was started in 2008, in an effort to characterize the microbial communities inhabiting different parts of the human body and their relation to health and disease.
One of the main interests in the Human Microbiome Project, is the scalp and its microbes in relation to dandruff. A French research group, Clavaud et al. (2013) have found that there may be a relation between the microbes on the scalp and dandruff, which is one of the most common symptoms of seborrheic dermatitis: a condition of scaly skin and red dry patches of skin (Misery et al., 2013). While dandruff is very common in France (about 16% of French citizens have dandruff), it is a global issue (Misery et al., 2013).
Central question
Other dermal diseases like acne (Wang et al., 2014) have been associated with imbalances or differences in the microbiomes of healthy and afflicted individuals. Clavaud et al. want to know if the same can be said for the scalp. Do the microbial communities of scalps with dandruff have different community members than scalps with no dandruff?
To start their experiment, Clavaud et al had their research participants use a standard, neutral shampoo for three weeks before sampling, and they were not allowed to use any other products in their hair. This allowed researchers to look at the baseline microbial community of the scalp of clean hair without the confounding variables coming from hair products (Clavaud et al., 2013).
Evidence
They identified microbes by targeting the 16S sequence for bacteria and 28S-ITS sequence for fungi. The target fragments were amplified via polymerase chain reaction (PCR) and sequenced with Sanger sequencing. These sequences were compared to a database of known 16S and 28S-ITS of various bacteria and fungi (Clavaud et al., 2013). In their results, they found the majority of bacteria sequences were from the genera Propionibacteria (49%) and Staphylococci (40%). Propionibacteria and Staphylococci dominated both dandruff and healthy scalps, with 99% of those sequences being identified at the species level as Propionibacteria acnes (P. acnes) and Staphylococci epidermis (S. epidermis), respectively. So both the dandruff and healthy scalps had communities mostly comprised of S. epidermis and P. acnes.
Because the dandruff and healthy scalps were dominated by the same community members, Clavaud et al. used quantitative PCR (qPCR) to determine the abundance of sequences of each community member. The qPCR results showed that although the community members may not differ between dandruff and healthy scalps, their abundances do differ (Figure 1A). A healthy scalp is characterized with a majority of P. acnes accounting for 75% of the scalp community, with roughly a quarter of the community belonging to S. epidermis. The opposite was found in the dandruff scalps: S. epidermis being the majority as 60% of the community and P. acnes accounting for only 15%.
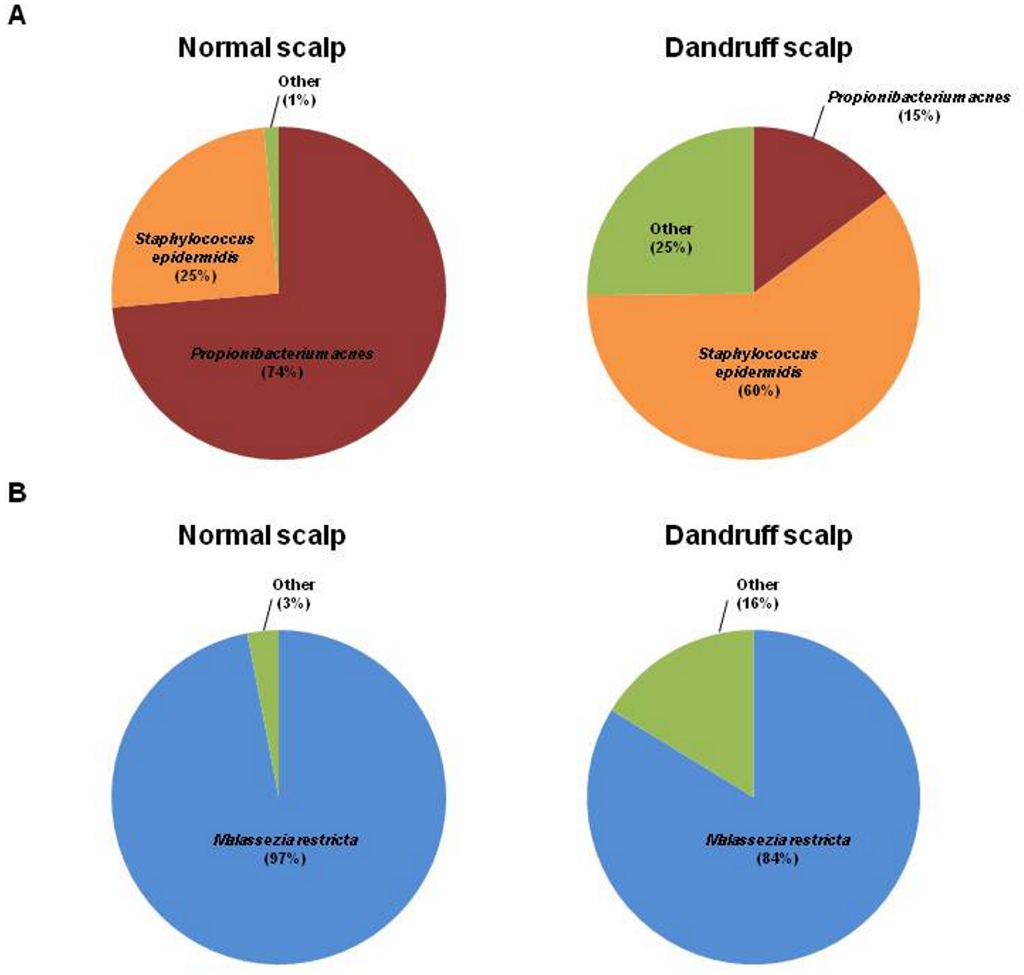
Roughly the same result was found in the fungal sequences. Malassezia restricta was the major fungal species present on the scalps with or without dandruff, but was more abundant in the healthy scalps than the dandruff scalps (2013).
These results are consistent with other studies investigating the relationships of P. acnes and S. epidermis in the skin. Wang et al. (2014) studied the balance of P. acnes and S. epidermis in relation to acne. They found that acne was related to a high proportion of P. acnes, which is known to prefer oily skin and ferment fatty substances, compared to other skin microbes. Wang et al. also found that S. epidermis was able to prevent P. acnes from growing (2014). In the case of acne, P. acnes may grow too large, ferment too much oil in our pores, and prevent the growth of other bacteria. In this case S. epidermis is preventing the growth of P. acnes and alleviating the disease.
The scalp provides a warm, oily environment for microbes to inhabit, and is similar to the microbiome of the face (Oh et al., 2016). However in the scalp, the roles of P. acnes and S. epidermis are switched. While fermenting fatty substances aggravated acne in oily skin, the fermentation of glycerol by P. acnes was shown to prevent colonization of pathogens in mice, including MRSA (Sanford & Gallo, 2013). P. acnes could be protecting the scalp from pathogens, while providing natural oils to hydrate the scalp and hair follicles. Thus, scalps may require larger proportions of P. acnes and smaller proportions of S. epidermis to be healthy. Reversing these proportions could lead to dandruff. If S. epidermis prevents the growth of P. acnes too much, there would not be as much glycerol fermentation, which means less oil for the scalp. Also, because S. epidermis can live in oily or dry conditions and P. acnes prefers oily skin, the dry scalp would not affect S. epidermis, but could hinder P. acnes (Sanford & Gallo, 2013). The dominance of S. epidermis could be amplifying the already dry conditions of the dandruff scalp, leading to the decline in P. acnes seen in the results of Clavaud et al.’s study (2013).
These results support the idea of dandruff being linked to an imbalance in the microbial communities, although Clavaud et al. remind us that these results only support a correlation, and not a causation. They would like to sample larger groups of people, and assess the interactions of microbes with scalp sebum production (2013). This study would also benefit by using next generation sequencing rather than Sanger sequencing. By using Sanger sequencing, the read depth of this study is small, about 200 sequences per sample (Clavaud et al. 2013), compared to next generation sequencing which can produce 50,000 reads on average for quality, microbial samples (Klindworth et al. 2013). A low reading depth glosses over rare species in the sample that could be affecting the scalp in a big way. Let’s say there is one pathogen that is known to cause itchy, rashy skin. If we only pick 5 microbes we may miss that pathogen, but we are more likely to sequence it if we pick 500 microbes. We should keep in mind that S. epidermis and P. acnes may not be the only microbes at play; there could be other key microbes that were not seen in this study because the reading depth was too low to catch these less abundant microbes.
My questions
In the end, Clavaud et al. could not conclude on the causation of dandruff, only the different compositions of dandruff and non-dandruff scalps. However, if future studies support the idea that the microbial community caused the diseased state, there could be probiotic treatments for dandruff that would fix the balance of the scalp’s microbiome. Like probiotic yogurt for your scalp. Of course this would depend on whether the environment caused the bacteria to change, or the bacteria cause the scalp to change. Do certain environmental factors like dry climate or hair product cause a scalp to develop dandruff, and this just happens to be a good habitat for Propionibacteria and Staphylococci to flourish? Or does the composition of the scalp microbiome cause dandruff?
To answer these questions, I think future studies of people in different countries and climates other than France would be valuable. Characterization of the scalp microbiome from climates different from France is necessary in order to measure any environmental effects. If all the studies had similar community results even with their varying climates, it could point the blame to the microbes instead of the environment. A metabolomics study would also be valuable. A metabolomics study analyses the proteins and other products produced by the microbes (Marchesi & Ravel, 2015). Then you could see if the microbes are producing a chemical that is harmful to our scalp, or if the healthy scalps have bacteria producing vitamins and oils that the dandruff scalp lacks.
Further reading
- For more on the Human Microbiome Project, click here and for the Human Microbiome Project Part 2, click here
- If you want to know more about seborrheic dermatitis, take a look at: Dessinioti, C., & Katsambas, A. 2013. Seborrheic dermatitis: Etiology, risk factors, and treatments:: Facts and controversies. Clinics In Dermatology, 31(4), 343-351. doi:10.1016/j.clindermatol.2013.01.001
- Here’s a similar paper on bacteria & fungal communities of scalps in China: Wang, L., Clavaud, C., Bar-Hen, A., Cui, M., Gao, J., Liu, Y., & … Breton, L. 2015. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Experimental Dermatology, 24(5), 398-400. doi:10.1111/exd.12684
References
- Clavaud, C., Jourdain, R., Bar-Hen, A., Tichit, M., Bouchier, C., Pouradier, F., & … Mouyna, I. 2013. Dandruff Is Associated with Disequilibrium in the Proportion of the Major Bacterial and Fungal Populations Colonizing the Scalp. Plos ONE, 8(3), 1-8. doi:10.1371/journal.pone.0058203
- Human Microbiome Project. https://hmpdacc.org/
- Human Microbiome Consortium. 2010. Structure and function of the healthy human microbiome. Nature 486, 207—214. doi:10.1038/nature11234
- Jincai, M., Kobayashi, D. Y. & Yee, N. 2009. Role of menaquinone biosynthesis genes in selenate reduction by Enterobacter cloacae SLD1a-1 and Escherichia coli K12. Environmental Microbiology, 11(1), 149-158. doi:10.1111/j.1462-2920.2008.01749.x
- Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M. & Glöckner, F.O. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi:10.1093/nar/gks808
- Marchesi, J.R. & Ravel, R. 2015. The vocabulary of microbiome research: a proposal. Microbiome, 3(31), 1-3. doi:10.1186/s40168-015-0094-5
- Misery, L., Rahhali, N., Duhamel, A., & Taieb, C. 2013. Epidemiology of Dandruff, Scalp Pruritus and Associated Symptoms. Acta Dermato-Venereologica, 93(1), 80-81. doi:10.2340/00015555-1315
- Oh, J., Byrd, A. L., Park, M., Kong, H. H., & Segre, J. A. 2016. Temporal Stability of the Human Skin Microbiome. Cell, 165(4), 854-866. doi:10.1016/j.cell.2016.04.008
- Sanford, J. A., & Gallo, R. L. 2013. Functions of the skin microbiota in health and disease. Seminars In Immunology, 25(5), 370-377. doi:10.1016/j.smim.2013.09.005
- Sender R, Fuchs S, Milo R. 2016. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14(8): e1002533. doi:10.1371/journal.pbio.1002533
- Tilg, H. & Kaser, A. 2011. Gut microbiome, obesity, and metabolic dysfunction. Journal of Clinical Investigation 121(6), 2126-2132. doi:10.1172/JCI58109
- Wang, Y., Kuo, S., Shu, M., Yu, J., Huang, S., Dai, A., & … Huang, C. 2014. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Applied Microbiology & Biotechnology, 98(1), 411-424. doi:10.1007/s00253-013-5394-8
