Background
Monogenic disorders are estimated to affect 1/100 people at birth (WHO). They are caused by individual mutations in individual genes that result in non-functional products such as RNA molecules or proteins. In the case of cystic fibrosis (CF), a mutation in an ion-transport protein causes CF patients to have thick, sticky mucus that easily traps bacteria, viruses and other contaminants that can cause disease. Thick mucus is particularly an issue in the lungs where trapped microbes can lead to lung inflammation and infection (Cystic Fibrosis Foundation). In healthy individuals, microbes are not uncommon in the upper and lower respiratory tracts. Usually, microbes are eliminated by little hairs that sweep microbes trapped in free-flowing mucus up and out of the respiratory tract to be coughed up or swallowed (Huffnagle et al., 2016). However, in CF patients thick mucus limits this elimination of microbes, trapping them in the lungs and respiratory tract (Fig. 1).
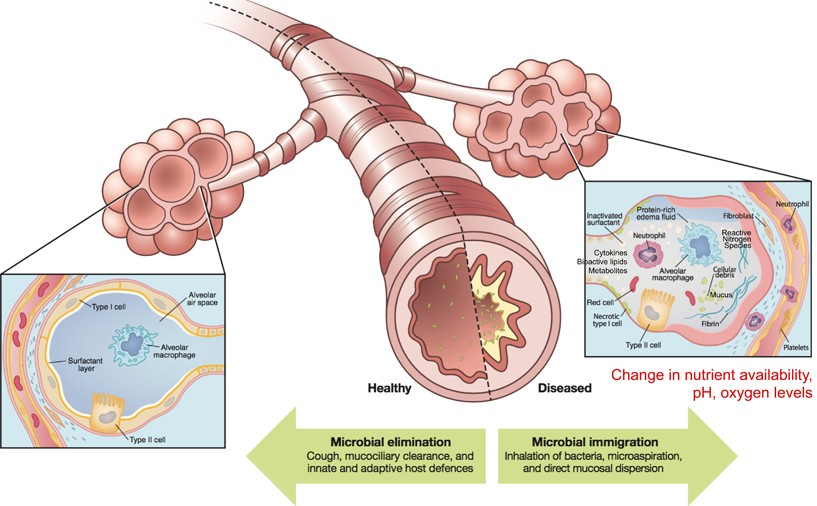
Often these microbes are bacteria that are able to grow and proliferate in the lungs of CF patients causing recurring infection and disease. These bacterial lung infections are one of the leading causes of death in CF patients (Mayo Clinic), so it is important to understand the relationships between the members of the lung microbial community, also known as the lung microbiota, and the human hosting them. With a greater understanding of these relationships, it might be possible to use the characteristics of a person’s lung microbiota to aid in the diagnosis and treatment of CF.
What is diversity and dominance in the microbiome?
In order to understand how the microbiome can change, let us grasp a better understanding of microbial diversity and dominance. These two factors are often used by microbiome researchers to get a baseline understanding of the changes occurring in a microbial community of a given environment (AKA the microbiome). In this case, the microbiome is the lungs of CF patients and the microbial community associated with them. The diversity of the microbiome is a measure of how many different microbes are present. Dominance, on the other hand, takes into account the abundance of the most common taxa. If the most abundant taxa make up a large portion of the total number of individual microbes, then that sample is said to have high dominance. However, if the most abundant microbial species only make up a small fraction of the total number of individuals, then that sample has low dominance. Using diversity and dominance, researchers can characterize general changes in the microbiome linked to variables like changes in disease state.
Central Question
How does the lung microbiome of cystic fibrosis patients change with decreasing lung function? In a new study, researchers from the United-Kingdom, Australia and the United-States asked how microbial diversity, abundance and composition changes with lung function (Cuthbertson et al., 2020).
Evidence
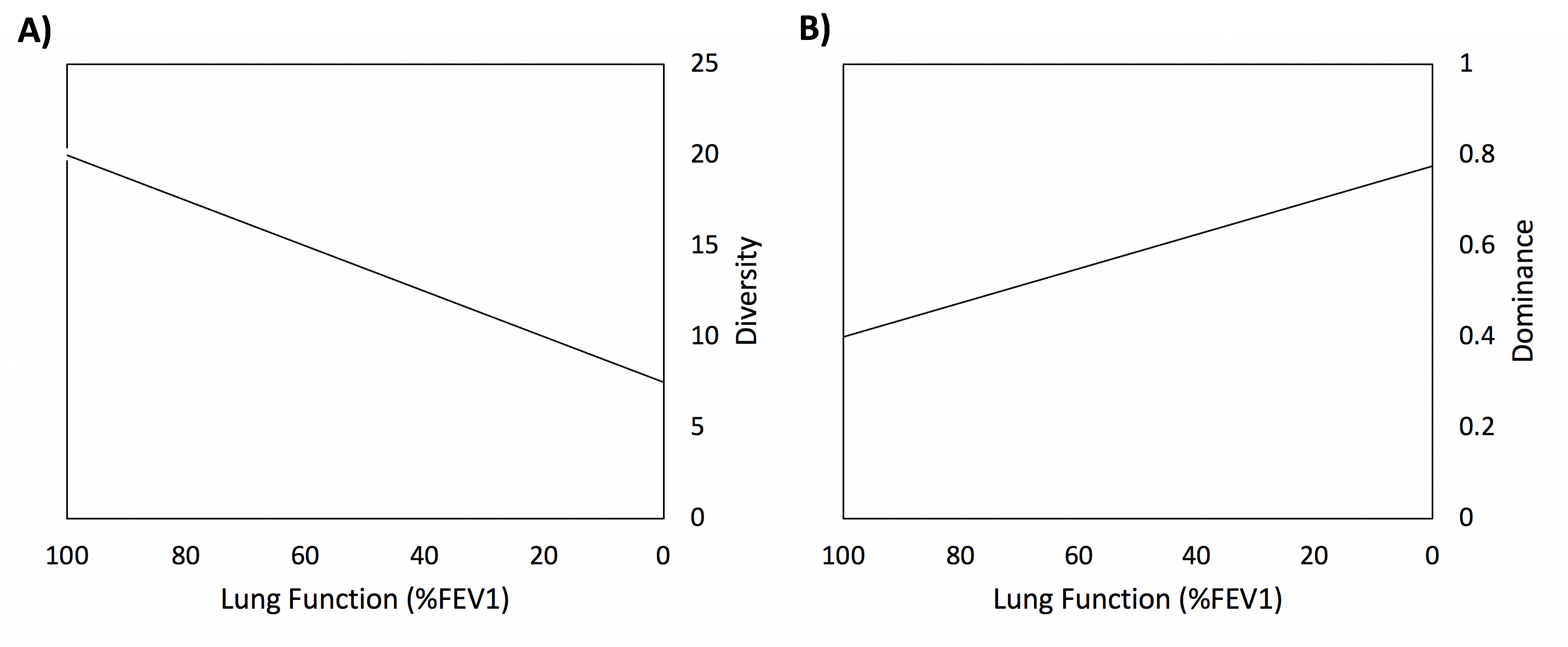
The researchers used sputum samples (the thick mucus coughed up from the lungs) to test for microbes from 299 individuals across 13 CF centers in the USA and Europe. They also used the amount of air exhaled in one second (FEV1) as an indicator of lung function. Using these methods, the authors found that decreases in microbial diversity and an increase in the dominance of CF associated pathogens were correlated with a decrease in lung function (Fig. 2). Could this mean that a decrease in microbial diversity and an increase in dominance could lead to reduced lung function? Not quite. The authors found that microbial diversity and dominance are correlated to lung function, but they did not provide evidence showing that one causes the other. Future research is needed to determine the direction of this relationship: whether changes in the microbiota cause decreases in lung function or decreases in lung function cause the microbiota to change.
Core vs. Satellite Taxa
In many cases, the composition of the lung microbiota is dominated by a few species. These species are part of the “core” microbiota which are present in relatively high abundances across many individuals and are often the primary focus of antibiotic treatment (Beck et al., 2012; Cuthbertson et al. 2020). “Satellite” taxa, on the other hand, make up a relatively low abundance both within individuals and across samples. In the second part of the study, the researchers separated the 598 different taxa they identified into either the core or satellite group. Individuals’ microbiota based on lung dysfunction into mild, moderate and severe lung function groups. When they reanalyzed the data, they found that decreases in diversity and increases in dominance were primarily only significant for the core taxa. Four of these core taxa included known CF pathogens (Elborn, 2013) present in relatively high abundances, such as Pseudomonas aeruginosa and Staphylococcus aureus which increased in relative abundance, and thus dominance, with increasing disease severity. This result is unsurprising given that reduced lung function is commonly caused by disease/infection in CF patients (Cystic Fibrosis Trust).
Why does the lung microbiota change with lung function?
Although this study was purely correlative, the researchers performed analyses to determine the potential contribution of significant variables towards the changes in the lung microbiota. They found that the use of antibiotics such as Flucloaxin and Meropenem, in addition to lung function, was the most significant contributing factor for changes seen in the core taxa, satellite taxa and overall microbiota. Interestingly, only the satellite taxa were significantly affected by geographical distance, hinting that only the microbes in low abundance change depending on where a patient lives. This difference between core and satellite taxa could indicate that it would be important to focus on the core taxa if using the lung microbiome as a tool in the diagnosis and treatment of infections in cystic fibrosis patients.
Future research and my questions
The results from this baseline study on the trends of the lung microbiota of cystic fibrosis patients provide a great foundation for future studies to determine the nature of the relationship between the lung microbiota and lung function. Future research questions could include the exploration of the metabolic pathways commonly present among lung microbiota and their correlation with lung function in cystic fibrosis patients. Researchers could collect metatranscriptome data from the microbes in the lungs of cystic fibrosis patients to analyze the types of proteins produced and how they affect lung function. Analyzing changes in the lung microbiome with antibiotic treatment could be another way to determine how lung microbiota could be involved in lung function. With more information, could the lung microbiome be a useful tool in the diagnosis and treatment of lung disease in cystic fibrosis patients? If the lung microbiota plays a large role in the manifestation and development of disease and infection, could altering the lung microbiome via probiotics be a useful form of treatment? These questions remain to be answered, but with more data, the applications of the lung microbiome to improve the lives of cystic fibrosis patients is certainly a possibility.
Further Reading
An overview of the study discussed above watch this abstract video: Cuthbertson et al., 2020
More information on the use of FEV1 in the assessment of lung function: Cirno and Marcin, 2018
For more information on one of the most common pathogens affecting CF patients, Pseudomonas aeruginosa: Bjarnsholt et al., 2010
References
Bjarnsholt T, et al. (2010) Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 5: 1-10.
Beck JM, Young VB, and Huffnagle GB (2012) The microbiome of the lung. Translational Research 160: 258-266.
Cirno E, and Marcin J (2018) FEV1 and COPD: how to interpret your results. Healthline n.p.
Cuthbertson L, Walker AW, Oliver AE et al. (2020) Lung function and microbiota diversity in cystic fibrosis. Microbiome 8: 45.
Cystic Fibrosis Foundation (n.d.) About Cystic Fibrosis. n.p. Retrieved: September 24, 2020.
Cystic Fibrosis Trust (n.d.) Lungs and Cystic Fibrosis. n.p. Retrieved: October 10, 2020.
Elborn, J.S. (2013) Current Approaches to the Management of Infection in Cystic Fibrosis. Current Pediatrics Reports 1: 141-148.
Huffnagle GB, Dickinson RP, and Lukacs NW (2016) The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunology 10: 299-306.
Mayo Clinic (n.d) Cystic Fibrosis. n.p. Retrieved: October 11, 2020.
WHO (n.d.) Genes and human diseases. Human Genomics in Global Health n.p. Retrieved: September 24, 2020.
